Abstract
To identify animals susceptible to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection or to determine whether SARS-CoV-2 contaminated meat is from a SARS-CoV-2-infected animal, a convenient and safe method was developed for rapid detection of SARS-CoV-2 in a replicating or non-replicating status in samples using reverse transcriptase–polymerase chain reaction (RT-PCR). This strategy can also be applied to develop assays for the detection of other viruses, either replicating or not.
Keywords: SARS-CoV-2, Negative-sense RNA, RT-PCR
Searching for animals susceptible to SARS-CoV-2 infection is one of the essential strategies to trace the origin of SARS-CoV-2 (Shi et al., 2020). It is also important to determine whether SARS-CoV-2-contaminated meat is from a SARS-CoV-2 infected animal or not. The established method to determine the status of SARS-CoV-2 replication in tissues or cells is through culturing these samples in a biosafety level 3 facility, however, this is very labor- and time-consuming, and unsafe for researchers/technicians.
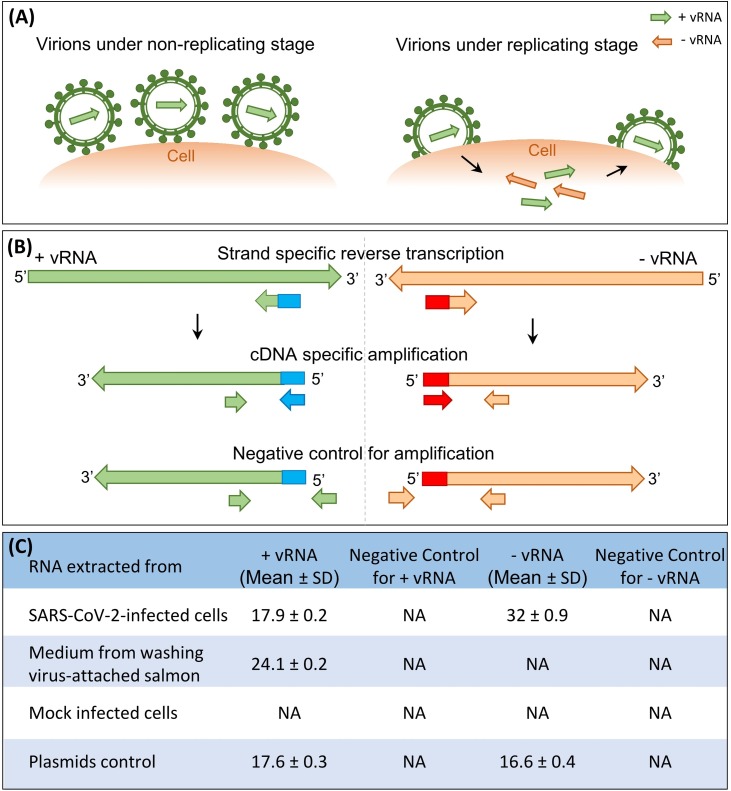
SARS-CoV-2 is a single-stranded positive-sense RNA virus. The viral negative-sense RNA is produced only when it is replicating in cells (Baltimore, 1971) (Figure 1 A). At present, most methods detect the total RNA of the virus, but the results do not indicate whether the virus is replicating, or not (Kim et al., 2020, WHO, 2020). Here, we designed and developed a simple RT-PCR assay to detect both viral positive- and negative-sense RNA simultaneously to determine whether the virus in tissues or cells is replicating, or not (Figure 1B).
Figure 1.
Detection of SARS-CoV-2 positive- and negative-sense RNA in a sample. (a) Illustration of replicating and non-replicating SARS-CoV-2. The viral negative-sense RNA was produced during the virus replicating stage in cells. (b) A schematic of the methodology of strand-specific RT-PCR. The blue and red fragments represent two different internal amplification controls. (c) The results of RT-PCR detection in different samples. NA means no specific PCR products amplified.
In brief, Vero-E6 cells were infected with SARS-CoV-2 and incubated at 37 °C for 48 h. The infected cells were then collected. Another sample consisting of the medium obtained after washing SARS-CoV-2-attached salmon was collected as previously described and is also explained in the supplement (Dai et al., 2020). Then the total RNAs were extracted from each sample for reverse transcription with the strand-specific primers (Figure 1B) (Deer et al., 2010). The results indicated that both viral positive- and negative-sense RNA were detected from the virus-infected cells, while only positive-sense RNA was detected from the medium, as shown in Figure 1C (see Supplemental Materials).
In summary, this assay to detect replicating SARS-CoV-2 in cell or tissue samples is convenient, rapid and safe for researchers/technicians. This strategy can also be applied to develop assays for the detection of other viruses, either replicating or not.
Author contributions
CP and ML conceived the idea and supervised the study; CP, JW, MD, HL, YN, and RY designed and performed the experiments; CP and ML wrote the manuscript.
Conflictof interest
The authors have applied for a Chinese patent based on the methods described in this study (Application No. CN202010994889.5).
Funding source
This work was supported by the Key Research and Development Project of Guangdong Province (202020012624900001).
Ethical approval
This work was approved by the National Health Commission of the People's Republic of China and performed in biosafety level 3 laboratory in South China Agricultural University.
Acknowledgements
We thank Dr Zhonghua Liu from Jiangsu Bioperfectus Technologies Co., Ltd. for his technical assistance.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.ijid.2021.01.043.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35(3):235–241. doi: 10.1128/br.35.3.235-241.1971. PMID: 4329869; PMCID: PMC378387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Li H., Yan N., Huang J., Zhao L., Xu S. Long-term survival of SARS-CoV-2 on salmon as a source for international transmission. J Infect Dis. 2020;(November) doi: 10.1093/infdis/jiaa712. jiaa712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deer D.M., Lampel K.A., González-Escalona N.G. A versatile internal control for use as DNA in real-time PCR and as RNA in real-time reverse transcription PCR assays. Lett Appl Microbiol. 2010;50(4):366–372. doi: 10.1111/j.1472-765X.2010.02804.x. [DOI] [PubMed] [Google Scholar]
- Kim J.M., Kim H.M., Lee E.J., Jo H.J., Yoon Y., Lee N.J. Detection and isolation of SARS-CoV-2 in serum, urine, and stool specimens of COVID-19 patients from the Republic of Korea. Osong Public Health Res Perspect. 2020;11(3):112–117. doi: 10.24171/j.phrp.2020.11.3.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368(6494):1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2020. Coronavirus Disease (COVID-19) Pandemic–Emergency Use Listing Procedure (EUL) Open for in Vitro Diagnostics.https://www.who.int/diagnostics_laboratory/EUL/en/ URL: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.