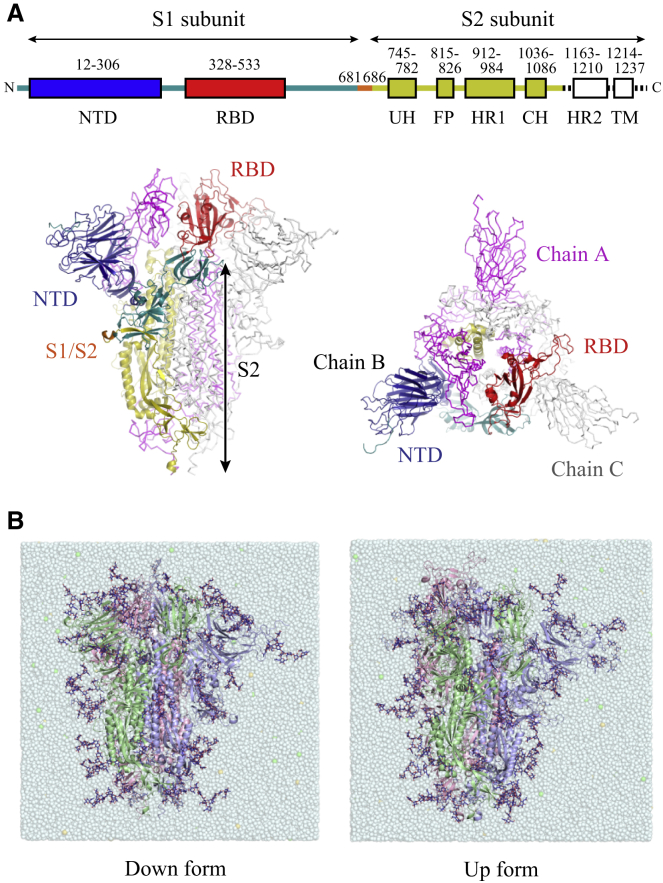
Figure 1.
The architecture and simulation models of SARS-CoV-2 S-protein. (A) The domain architecture of each monomer of a trimeric S-protein is shown (top). Here, the chains A and C are represented in magenta and gray, respectively. NTD, RBD, the rest of S1 subunit, and S2 except for HR2 and transmembrane domain in the chain B are highlighted in blue, red, green, and yellow (bottom). (B) Solvated simulation models of a fully glycosylated S-protein in down (left) and up (right) forms are shown. The chains A, B, and C are shown in red, green, and blue, respectively. The glycans are shown in blue stick representations. Throughout this study, RBD in chain A can take the up form in MD simulations and undergo the transition between down and up forms in TMD simulations. To see this figure in color, go online.