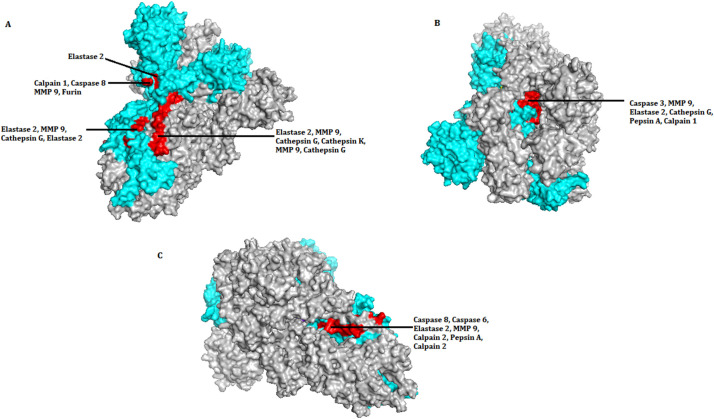
Fig. 2.
SARS-CoV-2 spike glycoprotein showing location of recognition sites for proteases from three orientations, A, B and C in both closed (6VXX) and open (6VYY) states. The positions are mutually exclusive. Out of the three chains in Spike protein structure, one chain was selected (marked in cyan) to mark the location of protease cleavage site. In the chain, red coloured patches indicate the presence of individual and overlapping positions of recognition sites. In orientation (A) proteolytic cleavage sites are shown in between amino acid sequence from 600 to 720. In orientations (B) and (C) cleavage sites are shown in between amino acid sequences 738–761 and 776 to 798 respectively. Only the sites accessible to solvent have been shown. Three sites that were inaccessible have not been shown (see Table 2).