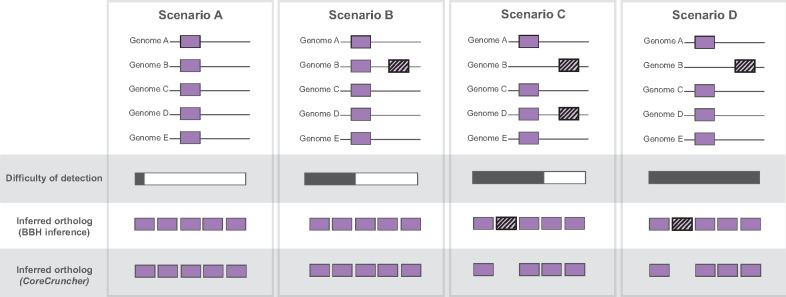
Fig. 1.
Impact of paralogs and xenologs on the inference of gene orthology. Scenario (A): No paralogs/xenologs. The orthologous gene is present in all the genomes; no paralogs or xenologs are present. Scenario (B): Within-paralogs/xenologs. The orthologous gene is present in all the genomes; one or more within-paralog/xenolog sequences are present. Scenario (C): Partially hidden paralogs/xenologs. The orthologous gene is missing in some genomes; some genomes are missing the orthologous sequence but contain a paralogous or a xenologous sequence (hidden paralog/xenolog); other genomes contain both the orthologous sequence and the paralogous or xenologous sequence (within-paralog/xenolog). Scenario (D): Completely hidden paralogs/xenologs. Some genomes are missing the orthologous sequence but contain a paralogous or a xenologous sequence (hidden paralog/xenolog); no within-paralogs/xenologs are present in other genomes. Plain boxes represent orthologous sequences; striped boxes represent paralogous or xenologous sequences. Scenarios A and B are expected to yield straightforward core gene predictions by BBH-based methods and CoreCruncher. Scenarios C and D will likely lead to the inclusion of paralogous and xenologous sequences in the core genome constructed with BBH-based approaches.