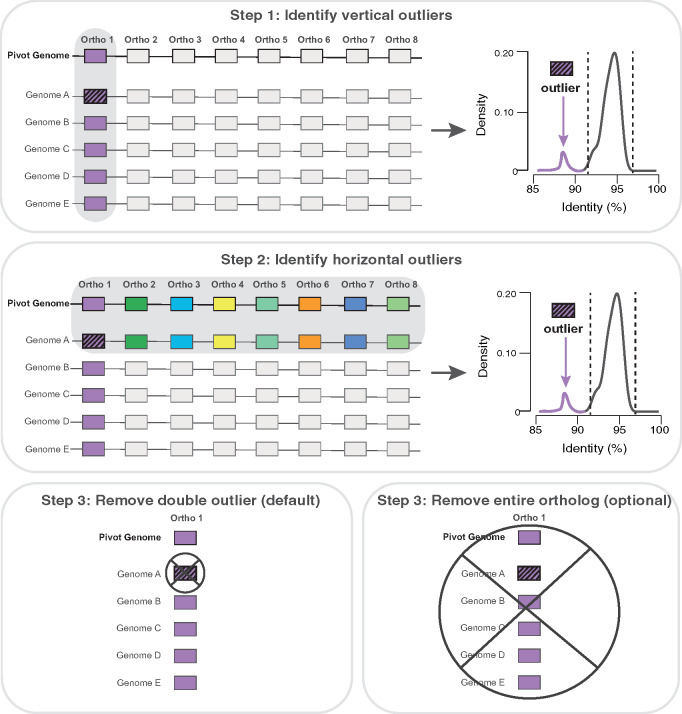
Fig. 2.
Identification of paralogs and xenologs with the double outlier test. CoreCruncher systematically tests for the presence of hidden paralogs/xenologs in each putative core gene. A sequence is inferred as paralogous/xenologous if it is identified as a vertical outlier and a horizontal outlier. Step 1. Vertical outliers: CoreCruncher builds distribution 1 for the putative core gene: the distribution of the identity scores of the best hit of each genome against the gene of the pivot genome. A sequence is considered as an outlier using Tukey’s fences: if its identity threshold is below Q1 − 1.5(Q3–Q1) or above Q3 + 1.5(Q3–Q1), with Q1 and Q3 the values of the first and third quartiles, respectively. Step 2. Horizontal outliers: sequences identified as outliers in step 1 are tested for the presence of horizontal outliers. CoreCruncher builds distribution 2 for the genome with a putative paralog/xenolog (i.e., an outlier was detected in step 1). The distribution is built by including all the putative orthologs of the genome with the putative paralog/xenolog against the pivot genome. The putative paralog/xenolog is considered a true paralog/xenolog if its identity score is also an outlier in distribution 2 using Tukey’s fences (see above). The paralog(s)/xenolog(s) inferred by the double outlier procedure is (are) then removed from the putative core gene. The putative core gene will be considered part of the core genome if present above the set frequency threshold used to define core genes (90% of genomes by default). When run with the stringent option, CoreCruncher will exclude any putative core gene with a paralog/xenolog identified with the double outlier test.