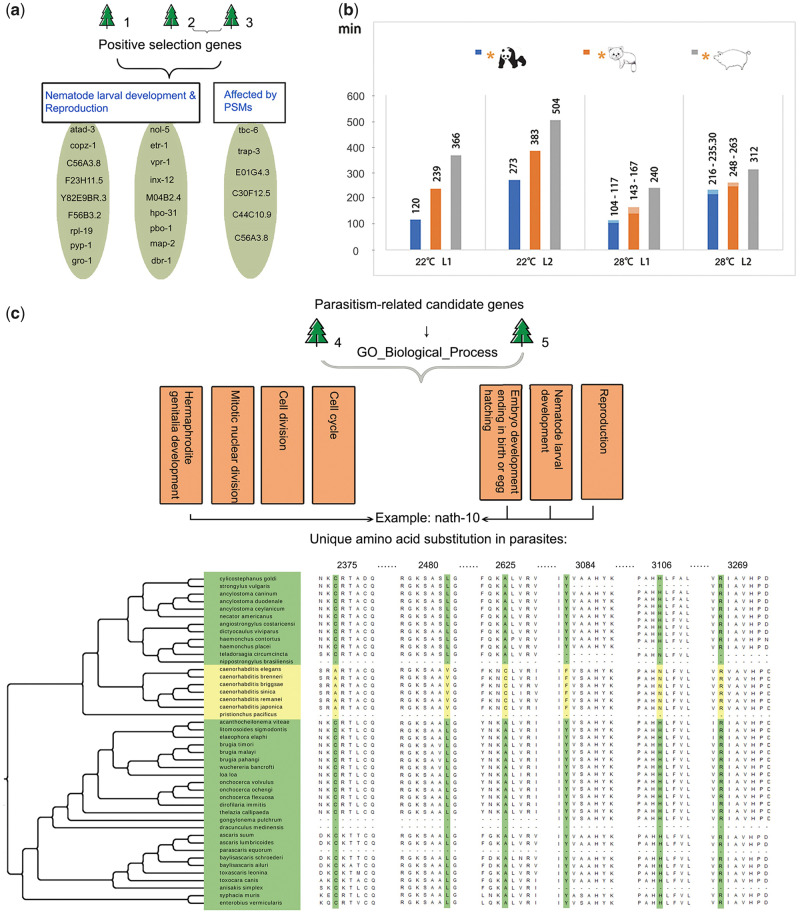
Fig. 2.
Positive selection related to the nutrient utilization and development of the roundworms from giant and red pandas, and positive selection and unique amino acid substitutions related to parasitism. (a) Two functional categories of positively selected genes in the roundworms from giant and red pandas. Trees 1, 2, and 3 indicate the first three positive selection analysis strategies, including both panda roundworms, only the giant panda roundworm, and only the red panda roundworm as the foreground branches, respectively. PSM, plant secondary metabolites. (b) In vitro development time of L1- and L2-stage larvae of three parasitic roundworms. The x-coordinate shows different temperature conditions and larval development stages, and the y-coordinate represents the development time (measured in minutes). A yellow asterisk indicates the host of the corresponding parasite. A lighter color in the column shows the fluctuation range of the development time. (c) GO biological process term enrichment of positively selected genes and unique amino acid substitutions related to parasitism. Trees 4 and 5 indicate the fourth and fifth positive selection analysis strategies, which both set all the parasitic roundworms as the foreground branches. Tree 4 only considered the positive selection of the most recent common ancestor lineage of five roundworms, and Tree 5 considered the positive selection of five roundworm lineages and their common ancestor lineages. Taking the nath-10 gene as an example, the arrow shows the enriched GO terms related to nath-10, and the multiple sequence alignments for 44 nematodes were ordered based on their phylogenetic relationships. Yellow represents free-living nematodes, and green represents parasites. Six amino acid substitutions were unique to all the parasites, with the number in the alignment being the location of unique amino acid substitutions.