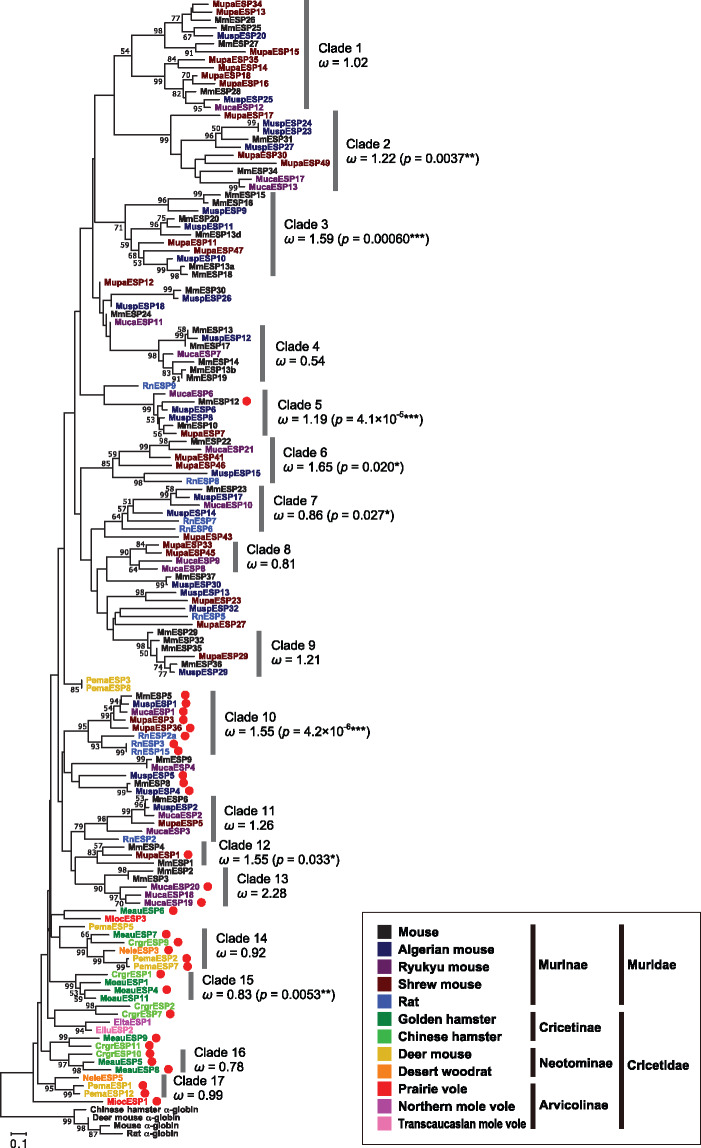
Fig. 5.
Neighbor-joining phylogenetic tree (Saitou and Nei 1987) constructed by MEGA7 (Kumar et al. 2016) for the amino acid sequences of 139 ESP mature sequences in 12 rodent species together with four α-globin genes used to determine the root. The evolutionary distances were computed using the Poisson correction method after all of the alignment gaps were eliminated for each sequence pair. Each sequence name was colored according to the color code for the species. The red filled circle represents a long (≥120 amino acids) mature sequence. Bootstrap values obtained from 500 replicates are shown for nodes with >50% bootstrap supports. Phylogenetic clades each of which is supported with a > 50% bootstrap value and contains three or more genes is numbered 1–17. The overall ω value calculated by PAML (Yang 2007) under model M0 is presented for each clade. The P value of the χ2 test for rejecting the null model of no positive selection (M7) over the alternative model of positive selection (M8) is also shown in parentheses for P < 0.05.