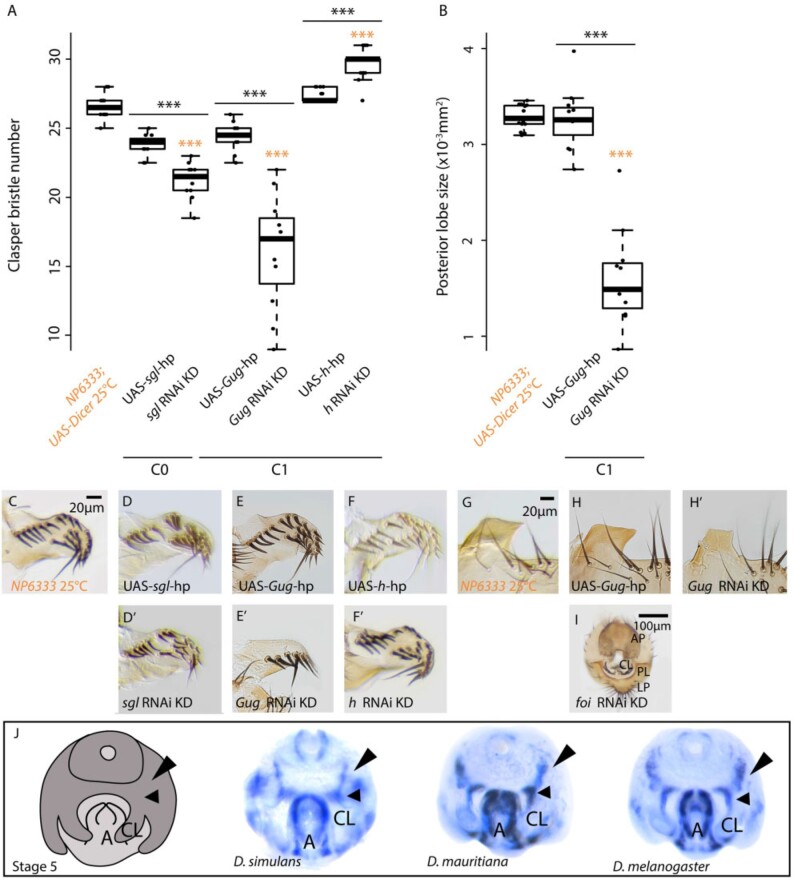
Fig. 2.
Functional analysis of positional candidate genes in D. melanogaster male genitalia. (A) Knocking down C0 candidate gene sgl (Tanaka et al. 2015) and C1 candidate gene Gug resulted in significantly fewer clasper bristles compared with both the UAS-Gug-hp (black asterisks, P < 0.001) and NP6333 driver controls (orange asterisks, P < 0.001). In contrast, knocking down C1 candidate gene h resulted in significantly more clasper bristles compared with the NP6333 driver (P < 0.001, orange asterisks) and UAS-h-hp controls (P < 0.001, black lines and black asterisks). (B) In addition, knocking down C1 candidate gene Gug resulted in the development of significantly smaller posterior lobes compared with the UAS-Gug-hp (orange asterisks) and NP6333 driver controls (P < 0.001, black asterisks). Asterisks indicate significant differences detected with Tukey’s pairwise comparisons, where P < 0.001*** and P > 0.05 = “ns” (supplementary file 6, Supplementary Material online). Orange indicates comparisons between the NP6333 driver control and UAS-gene-hp controls/gene knockdowns, whereas comparisons between UAS controls and knockdowns are indicated by black lines and black asterisks. Boxes indicate the range, upper and lower quartiles, and median for each sample. hp, hairpin; KD, knockdown. (C–I) Morphology of claspers and posterior lobes in NP6333 driver controls, UAS controls, and gene knockdowns (D–F′ and H, H′). (J) An illustration of stage 5 male genitalia (excluding the posterior lobes) and in situ hybridizations of Cpr66D in Dsim w501, Dmau D1, and Dmel w1118. Cpr66D transcripts were detected in a wider domain along the clasper inner edge (small arrowheads) and in bands extending toward the anal plates (large arrowheads) in the two species with larger clasper. Crpr66D is also expressed in the aedeagus of all three species. CL, clasper primordia; A, aedeagus (internal genitalia). Note that sgl RNAi knockdown data were generated in Tanaka et al. (2015) and reanalysed here.