Abstract
The adaptive radiations of East African cichlid fish in the Great Lakes Victoria, Malawi, and Tanganyika are well known for their diversity and repeatedly evolved phenotypes. Convergent evolution of melanic horizontal stripes has been linked to a single locus harboring the gene agouti-related peptide 2 (agrp2). However, where and when the causal variants underlying this trait evolved and how they drove phenotypic divergence remained unknown. To test the alternative hypotheses of standing genetic variation versus de novo mutations (independently originating in each radiation), we searched for shared signals of genomic divergence at the agrp2 locus. Although we discovered similar signatures of differentiation at the locus level, the haplotypes associated with stripe patterns are surprisingly different. In Lake Malawi, the highest associated alleles are located within and close to the 5′ untranslated region of agrp2 and likely evolved through recent de novo mutations. In the younger Lake Victoria radiation, stripes are associated with two intronic regions overlapping with a previously reported cis-regulatory interval. The origin of these segregating haplotypes predates the Lake Victoria radiation because they are also found in more basal riverine and Lake Kivu species. This suggests that both segregating haplotypes were present as standing genetic variation at the onset of the Lake Victoria adaptive radiation with its more than 500 species and drove phenotypic divergence within the species flock. Therefore, both new (Lake Malawi) and ancient (Lake Victoria) allelic variation at the same locus fueled rapid and convergent phenotypic evolution.
Keywords: standing genetic variation, cichlid fishes, convergent evolution, color patterns, evolutionary genomics, adaptive radiations
Introduction
Understanding how genetic variation translates into phenotypic diversity is an important goal in evolutionary biology. Repeatedly evolved phenotypes are particularly interesting for the study of the genetic basis of phenotypic diversity because they provide natural replicates that can inform whether the same evolutionary mechanisms have recurrently generated these phenotypes (Kuraku and Meyer 2008; Protas and Patel 2008; Stern 2013; Elmer and Meyer 2011;Elmer et al. 2014; Kratochwil and Meyer 2015; Kratochwil et al. 2018). Repeated evolution can result either from evolution through independent de novo mutations occurring in different species; or from preexisting variation that can be recruited via introgression or from standing genetic variation in a common ancestor (Stern 2013). Most de novo mutations are expected to be neutral or deleterious (Ohta 1992), whereas old standing genetic variation has likely already been purged from deleterious alleles due to previous selection. Adaptation from standing genetic variation is generally thought to be faster, as alleles reach fixation more quickly. Thereby, standing genetic variation might facilitate rapid diversification (Barrett and Schluter 2008; Marques et al. 2019). Ancestral variants can be more easily reassembled into new combinations, whereas fixation of de novo mutations is predicted to result in a slower diversification process (Barrett and Schluter 2008; Hedrick 2013; Marques et al. 2019). Accordingly, recent studies recognized the recruitment of alleles from standing genetic variation as an important evolutionary mechanism in driving the rapid phenotypic diversification found in adaptive radiations (Colosimo et al. 2004; Hines et al. 2011; Seehausen 2015; Lamichhaney et al. 2016; Han et al. 2017; Meier, Marques, et al. 2017; Bassham et al. 2018; Malinsky et al. 2018; Nelson and Cresko 2018; Salzburger 2018; York et al. 2018; Lewis et al. 2019; Svardal et al. 2020; Kautt et al., forthcoming). However, with a few exceptions (Colosimo et al. 2004; Hines et al. 2011; Lamichhaney et al. 2016; Meier, Marques, et al. 2017; Lewis et al. 2019), most studies reporting evidence for the importance of ancestral standing genetic variation across whole genomes lacked knowledge of genotype–phenotype connections (Malinsky et al. 2018; Nelson and Cresko 2018; Svardal et al. 2020). As a consequence, the specific impact of old genetic variation on phenotypic diversification often remains elusive.
The adaptive radiations of cichlid fishes offer a great opportunity to investigate the contribution of standing genetic variation to rapid adaptive divergence, due to their exceptionally high diversity in species and the repeated evolution of multiple phenotypes (Meyer et al. 1990; Meyer 1993; Stiassny and Meyer 1999; Kocher 2004; Genner and Turner 2005; Henning and Meyer 2014). Within the Great Lakes of the African Rift Valley, cichlids diversified into hundreds of endemic species in several lakes of different sizes and ages. In the three East African Great Lakes alone—Lake Victoria, Lake Tanganyika, and Lake Malawi—more than 1,200 cichlid species evolved (Salzburger and Meyer 2004). Recent studies showed that the onset of the exceptionally rapid adaptive radiation in Lake Victoria, in which at least 500 species evolved within the past 15,000 years (Johnson et al. 2000; Verheyen et al. 2003; Wagner et al. 2013), was fueled by high levels of genome-wide standing genetic variation (Seehausen 2004; Meier, Marques, et al. 2017). The Lake Victoria cichlid flock is derived from divergent lineages of the geologically older Lake Kivu (Verheyen et al. 2003) and adjacent rivers (Salzburger et al. 2005; Meier, Marques, et al. 2017), which started diversifying about 100–200 ky ago (Verheyen et al. 2003; Seehausen 2006; Genner et al. 2007). The older radiation of Lake Malawi cichlids encompasses about 700 species (Turner et al. 2008), which are believed to have evolved within the last 800 ky (Meyer et al. 1990; Danley and Kocher 2001; Brawand et al. 2014). Recently, whole-genome resequencing revealed that standing genetic variation contributed to the high diversification rates of this adaptive radiation (Svardal et al. 2020). Furthermore, standing genetic variation derived from ancestral lineages was also reported for Lake Tanganyika cichlids (Irisarri et al. 2018), the oldest and phenotypically most diverse of the three East African cichlid fish adaptive radiations (Sturmbauer and Meyer 1992; Salzburger et al. 2005; Koblmüller et al. 2008).
Within and between these different adaptive radiations multiple phenotypes have evolved repeatedly (Stiassny and Meyer 1999). This is exemplified by melanic horizontal stripes, an adaptive phenotype that is often associated with shoaling behavior and a piscivorous feeding mode (Seehausen and Alphen 2001). Previous work identified the gene agouti-related peptide 2 (agrp2, also called asip2b) as a major effect locus for stripe pattern divergence in African cichlids (Kratochwil et al. 2018). The teleost-specific gene agrp2/asip2b and its paralogs have been previously associated with pigmentation phenotypes (Zhang et al. 2010; Manceau et al. 2011; Ceinos et al. 2015). In zebrafish, agrp2 is mainly expressed in the pineal gland. Biochemically it acts as an antagonist of melanocortin receptors (Zhang et al. 2010). In cichlids, agrp2 has been demonstrated to also have a function in the skin, where it controls the presence of stripe patterns. High expression of agrp2 inhibits stripe patterns, while low expression permits their development. Yet, prior work could not identify the exact causal haplotypes and their evolutionary origin(s). The adaptive importance of horizontal stripes (Seehausen and Alphen 2001), together with a well-resolved genotype-to-phenotype connection (Kratochwil et al. 2018), makes the agrp2 locus an ideal target to investigate the role of preexisting standing genetic variation versus de novo mutations for driving adaptive phenotypic divergence in rapidly evolving species flocks.
Here, we addressed whether striped and nonstriped fish of the parallel and independent radiations of Lake Victoria, Lake Malawi, and Lake Tanganyika show the same signals of genomic divergence explaining the convergent evolution of stripe patterns. By including genomic sequences of more basal lineages of the Lake Victoria radiation from Lake Kivu and adjacent rivers we furthermore traced back the evolutionary origin of the causal major effect allelic variants of Lake Victoria cichlids.
Results and Discussion
Stripe Pattern Convergence and Diversification in African Cichlid Fish Radiations
To reconstruct the evolutionary history of the haplotypes associated with stripe patterns, we investigated the genomic interval around the agrp2 gene with a combination of target enrichment (∼30-kb agrp2 region ± 100 kb) and whole-genome resequencing as the agrp2 locus was previously shown to be associated with horizontal stripes in cichlids of the three African Great Lakes (Henning et al. 2014; Kratochwil et al. 2018). Data were collected from 213 individuals from the three great African species flocks (number of individuals/species in Lake Malawi n = 143/111, Lake Tanganyika n = 26/23, Lake Victoria n = 36/22; supplementary table S1, Supplementary Material online).
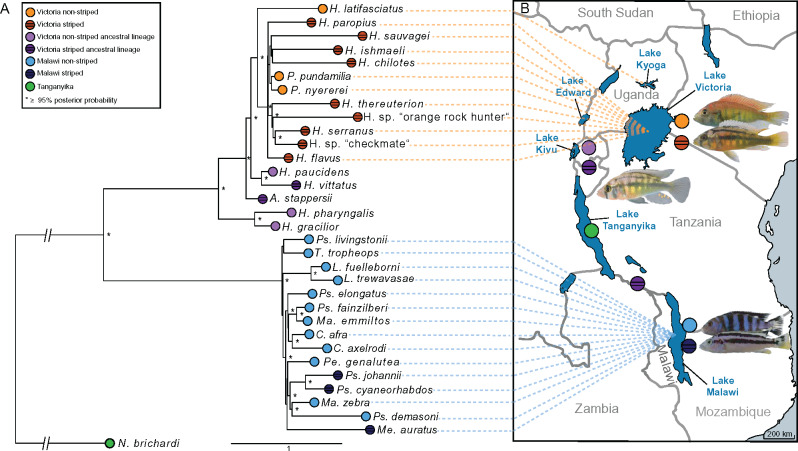
We inferred a species tree of the sampled species based on 6,545 genome-wide randomly selected loci of 3 kb from 33 high-quality genomes. The phylogeny from this state-of-the-art, high-density data set agrees with previous reports based on mitochondrial (Meyer et al. 1990), RAD-seq (Wagner et al. 2013) and, most recently, genomic data (Malinsky et al. 2018; Svardal et al. 2020). All phylogenies show strong discordance between the stripe phenotype and phylogeny showing that stripes clearly evolved repeatedly (fig. 1).
Fig. 1.
Recurrent evolution of horizontal stripes in East African cichlids. (A) Phylogeny of East African cichlid lineages based on 6,545 genome-wide random loci of 3 kb. Node color indicates lake of origin (Malawi in blue, Tanganyika in green, Victoria in orange, and the Lake Victoria outgroups in purple) and if the species shows horizontal stripes (dark color with stripes) or not (light color). (B) Map of the African Great Lakes and surrounding lakes with dotted lines connecting species in the phylogeny to their lake of origin. Horizontal stripes are present in about one-third of the species in all radiations. Photographs from top to bottom: Pundamilia nyererei, Haplochromis sauvagei, Astatotilapia stappersii, Pseudotropheus demasoni, Melanochromis kaskazini. Photograph credits: Jan Gerwin, Claudius Kratochwil, Adrian Indermauer, Sabine Urban.
Stripes in Lake Malawi and Victoria Radiations Are Associated with the Same Gene but Different Noncoding Regions
Using whole-genome resequencing and target enrichment data, we calculated relative genetic differentiation (FST) between striped and nonstriped species for the cichlid radiations of Lakes Tanganyika, Malawi, and Victoria over the 672,091 filtered biallelic single-nucleotide polymorphisms (SNPs) called across the whole ∼10-Mb scaffold 3 containing the agrp2 gene. Parallel evolution drives certain mutations to fixation independently in different populations and thereby acts on very local genomic regions. Therefore, we used the software Saguaro for FST calculation which implements an algorithm that sets out to identify and pinpoint such regions using a Hidden Markov Model and a Neural Network, applied in an interleaved fashion. Saguaro then infers local relationships among individuals in the form of genetic distance matrices and assigns segments across the genomes to these topologies.
In the Lake Tanganyika radiations, we did not find regions of elevated FST between striped and nonstriped species around agrp2 (supplementary fig. S1, Supplementary Material online), although a link between agrp2 expression and stripes has been shown earlier (Kratochwil et al. 2018).
The Lake Tanganyika species flock is more than 10 My old and consists of several ancient independent radiations (Salzburger et al. 2002, 2005; Clabaut et al. 2005; Koblmüller et al. 2008; Takahashi and Koblmüller 2011) with a complex history of repeated colonization events (Nishida 1991; Salzburger et al. 2002). Therefore, the missing association of alleles within the agrp2 locus with stripes is likely explained by more complex genetic mechanisms of stripe formation and might involve multiple cis-regulatory loci and/or trans-regulatory mechanisms as well as additional modifier loci.
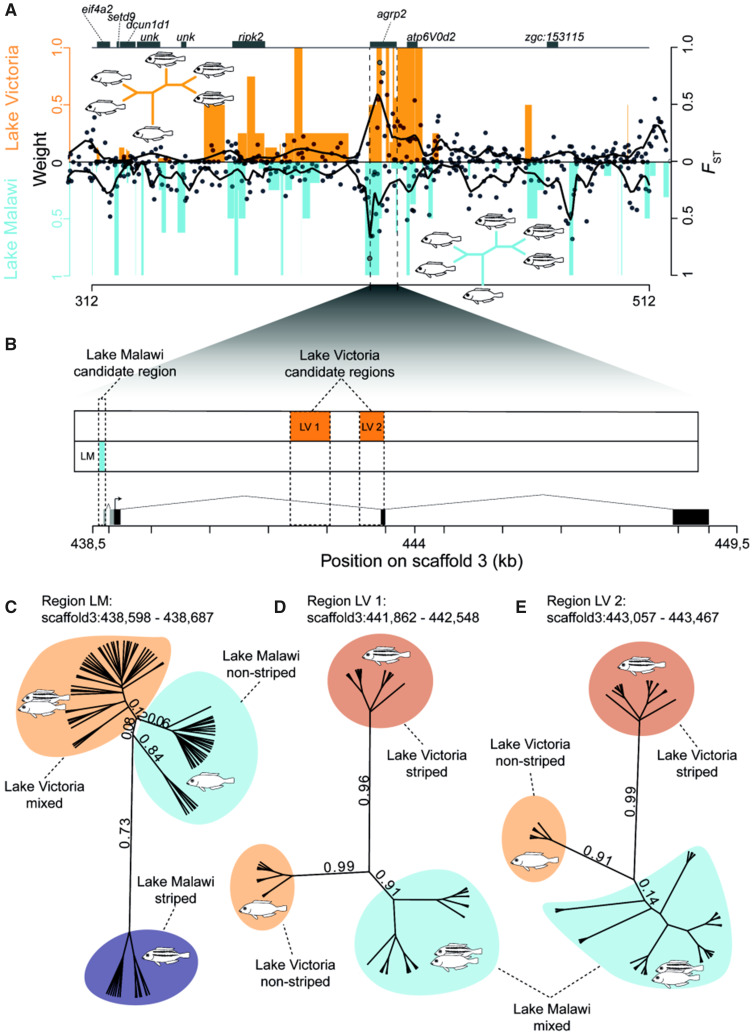
Both adaptive radiations of Lakes Victoria and Malawi are composed of a single lineage of haplochromine cichlids which evolved within the last 2–4 My in Lake Malawi and in Lake Victoria within 0.01–1 My (Meyer et al. 1990; Kocher 2004; Turner 2007; Brawand et al. 2014). Among the 700 endemic Lake Malawi cichlids, the agrp2 locus shows elevated differentiation among the littoral rock-dwelling mbuna which contains at least 200 species (fig. 2A) (Danley and Kocher 2001). Within this lineage, the strongest differentiation between striped and nonstriped species includes the 5′ untranslated region (UTR) of agrp2 (FST = 0.85 vs. scaffold mean 0.09; fig. 2A and B). In a gene tree inferred based on this region, the topology clearly separates striped mbuna from nonstriped mbuna but not the Lake Victoria phenotypes (fig. 2C). A single species, Petrotilapia nigra, is heterozygous for two of the three variants close to and within the 5′-UTR (supplementary fig. S2, Supplementary Material online) and shows a very indistinct stripe pattern. Variants within region LM are unique to striped Lake Malawi mbuna, and there is no association in nonmbuna or Lake Victoria cichlids (fig. 3). These variants, therefore, most likely constitute de novo mutations that evolved within the last ∼300 ky in the Lake Malawi mbuna radiation (Genner et al. 2007). However, this association between the agrp2 locus and stripes vanishes when comparing the whole Lake Malawi data set including nonmbuna species (supplementary fig. S1, Supplementary Material online).
Fig. 2.
The same gene but different lake-specific regulatory regions are associated with the repeated evolution of stripe patterns in the adaptive radiations of haplochromine cichlids of Lakes Malawi and Victoria. (A) Association of stripes with genomic regions. Black dots represent midpoints of every associated region (FST value) as identified by Saguaro and black lines are smoothened local regressions between striped and nonstriped species from Lake Victoria (top) and Lake Malawi mbuna species (bottom). This is plotted together with topology weights for topologies in which striped and nonstriped species are reciprocally monophyletic (orange bars Lake Victoria, blue bars Lake Malawi mbuna). Each value gives the proportionate contribution of a particular taxon tree to the full tree with values ranging from 0 to 1. An example for such a topology in which striped species are reciprocally monophyletic is provided for both radiations. (B) Gene structure of agrp2 with regions of elevated FST (FST > 0.75). Gray boxes indicate two isoforms of the 5′-UTR that harbors variants associated with stripe divergence in Lake Malawi mbuna cichlids. (C) Unrooted gene tree from the region of highest differentiation in Lake Malawi (LM, 90 bp). The two monophyletic groups of nonstriped Malawi cichlids differ in two variants at positions 438,598 and 438,657 (fig. 3 and supplementary fig. S2, Supplementary Material online). Numbers represent posterior probabilities. (D) Unrooted gene tree inferred from the region of highest differentiation in Lake Victoria (LV 1, 687 bp). (E) Unrooted gene tree from the region of second highest differentiation in Lake Victoria (LV 2, 411 bp).
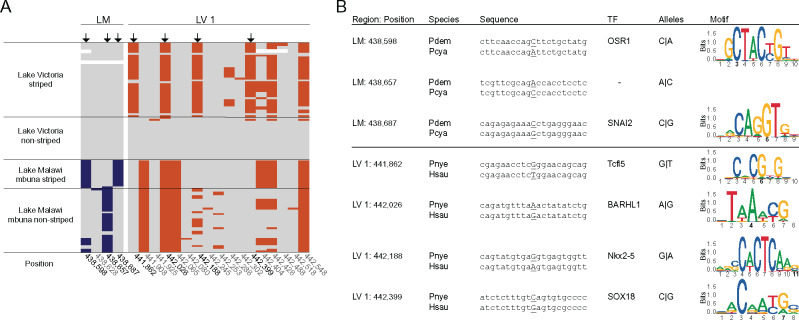
Fig. 3.
Different lake-specific substitutions are associated with stripe pattern evolution in Lakes Malawi and Victoria. (A) Gray represents the “nonstriped” allele of the Pundamilia nyererei reference genome. All nonreference alleles within candidate region LM are indicated in blue. Substitutions in LM are not shared between Lake Victoria and Lake Malawi cichlids suggesting that they originated de novo in Lake Malawi mbuna. Orange indicates substitutions that were found in candidate region LV 1 that are associated with stripe patterns in Lake Victoria cichlids. Although those haplotypes are divergent between striped and nonstriped cichlids in Lake Victoria, this is not the case in Lake Malawi mbuna. Black arrows highlight substitutions that are highly divergent between striped and nonstriped species (>90% frequency difference since some striped species are heterozygous, see also fig. 4A and supplementary fig. S3, Supplementary Material online). (B) A screen for potential TFBSs in a nonstriped (Pseudotropheus demasoni, Pdem, in Malawi and Pundamilia nyererei, Pnye, in Victoria) and striped (Pseudotropheus cyaneorhabdos, Pcya, in Malawi and Haplochromis sauvagei, Hsau, in Victoria) representative species reveals several TFBSs that are affected by stripe-associated alleles. Shown are those TFs with the highest delta relative score. Variable positions in the motif sequence are capitalized and underlined.
For the Lake Victoria radiation, the agrp2 locus was shown to be highly differentiated between striped and nonstriped species (fig. 2A). The two most differentiated regions (FST = 0.87 and FST = 0.78 vs. scaffold mean of 0.06) are directly upstream of the second exon and largely overlap (58,4% overlap) with a cis-regulatory active region (442,318–443,409) that was previously identified based on Sanger sequencing of three Lake Victoria species and experimentally tested using a transgenic reporter assay (Kratochwil et al. 2018). Taken together, the Lake Victoria regulatory interval (including both highly associated regions, LV 1 and LV 2; fig. 2B) has a size of ∼1.23 kb and is likely composed of several smaller cis-regulatory elements such as enhancers and/or silencers (fig. 2B). In contrast to the topology of the region LM, the gene trees inferred from these two regions of highest differentiation (LV 1 and LV 2) clearly separate striped from nonstriped Lake Victoria cichlids (fig. 2D and E). This pattern supports the hypothesis that different regulatory regions at the same locus facilitate convergent evolution of stripe patterns across different cichlid radiations.
To further support the association of the identified cis-regulatory intervals found in Lakes Malawi and Victoria with stripe patterns, we employed a second, complementary approach, in which we assessed topology weights with TWISST (Van Belleghem et al. 2017). The TWISST results strongly support a topology that groups species by stripe phenotype (fig. 2A). The adjacent gene, atp6V0d2, also exhibited pronounced topology grouping by stripe phenotype, but previous work did not reveal any fixed mis- or nonsense mutations or differential expression (Kratochwil et al. 2018).
Noncoding Variants Predict Changes in Transcription Factor Binding in Highly Divergent Regions of Both, Lakes Malawi and Victoria
Both highly divergent regions are noncoding and might therefore contribute to variation in agrp2 transcription and/or translation. The highly associated 90-bp region in Lake Malawi (LM; fig. 2B and C) overlaps with the 5′-UTR of agrp2. 5′-UTRs can contain transcription factor-binding sites (TFBSs) (Barrett et al. 2012; Lavallee-Adam et al. 2017) but also have been shown to play important roles in posttranscriptional regulation (Araujo et al. 2012) and could therefore lead to variation in transcript stability or translation rate. To provide additional evidence that the substitutions within and close to the 5′-UTR of agrp2 in Lake Malawi mbuna might influence agrp2 transcription, we screened these regions for potential TFBSs that are likely affected by the associated variants. For this, we used sequences from a representative nonstriped and striped species (nonstriped Pseudotropheus demasoni, and striped Ps. cyaneorhabdos) flanking the three variant sites within candidate region LM ±10 bp (fig. 3 and supplementary fig. S2 and table S2, Supplementary Material online). The sequence flanking the first variant (position 438,598) contained 18 TFBSs of which ten have a delta relative score of >0.1 (Materials and Methods, supplementary table S2 and fig. S2, Supplementary Material online) therefore suggesting a higher TF-binding affinity in the nonstriped (the species with high expression of the “stripe-repressor gene” agrp2) than in the striped species. The TFs include tfc3 that was associated with pigmentation previously (Dorsky et al. 2000). For the second variant, we did not identify TFBSs with a delta relative score of >0.1. For the third variant in LM (position 438,687), 18 TFBSs were predicted within the 5′-UTR and five of these show a delta relative score of >0.1. These five transcription factors (TFs) have all (snai2, two variants of tfap2e, tfap2a, and six1) been linked to pigmentation (Sanchez-Martin et al. 2002; Van Otterloo et al. 2010; Yang et al. 2019) and are expressed in the skin, melanophores, or neural crest cells in zebrafish (https://zfin.org/, last accessed September 25, 2020). The neural crest is a highly migratory population of embryonic cells from which melanophores originate (Le Douarin and Kalcheim 1999). In conclusion, variants within the 5′-UTR of agrp2 might have led to lower expression or transcript stability of agrp2. The resulting low expression of agrp2 might in turn have triggered the de novo appearance of the stripe phenotype in Lake Malawi mbuna cichlids.
The most highly associated region in the Lake Victoria radiation (also when we included closely related riverine and Lake Kivu lineages; fig. 4) is LV 1. We therefore also screened this region for potential TFBSs using the nonstriped species Pundamilia nyererei (Pnye) and striped Haplochromis sauvagei (Hsau). Our analysis revealed high delta relative scores for several TFs with a known function in pigmentation pathways. For example, tcfl5 at position 441,862 belongs to a group of TFs involved in the Wnt signaling pathway. In zebrafish, Wnt signaling activates nacre, a zebrafish homolog of mitf, a key regulator of pigment synthesis, which in turn leads to pigment cell differentiation. Position 442,188 harbors a TFBS for zeb1, which in cichlids represses the expression of mitf (Albertson et al. 2014). The sequence around variant position 442,399 has a TFBS for sox18. The sequence around position 442,399 contains six more TFBSs belonging to the sox family of TFs with lower delta relative scores. Sox proteins including Sox18 regulate and interact during all stages of the melanocyte/melanophore life cycle (Harris et al. 2010). Some TF-binding differences are shared between LM and LV (i.e., nfix, spi1, nr2c2(var.2), zeb1, rbpj, sox3, sox10) suggesting that transregulatory factors might be identical in both radiations whereas cis-regulatory elements are not.
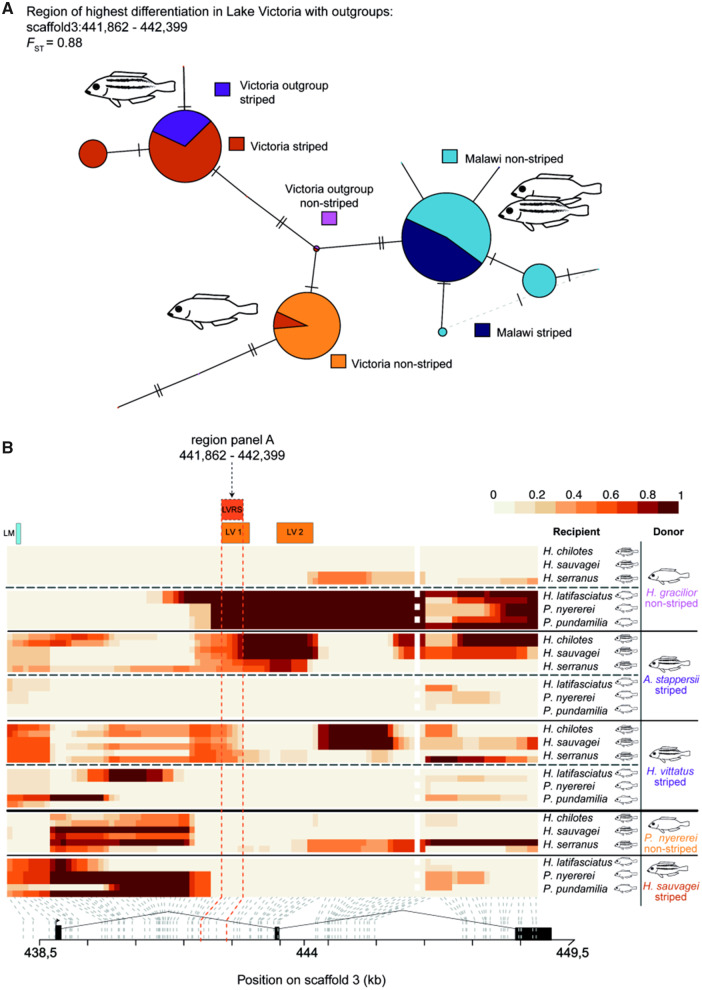
Fig. 4.
The evolutionary origin of the major effect haplotype of Lake Victoria stripe divergence predates the adaptive radiation of Lake Victoria. (A) Haplotype network of the highest FST region (LVRS, 538 bp) between striped and nonstriped species from the Lake Victoria superflock shows the stripe and nonstripe haplotype. Mismatches represent heterozygous individuals, that is, not all species that were assigned a stripe phenotype are homozygous for the stripe haplotype. Interestingly, these species do not show a complete stripe pattern consisting of a dorsolateral and a midlateral stripe but either show only one stripe (Haplochromis paropius) or represent a striped individual of a species (H. phythophagus) that displays a polymorphism by presence/absence of horizontal stripes. After FST calculation, we included the Lake Malawi mbuna which do not share the Lake Victoria stripe haplotype. However, the riverine Astatotilapia stappersii, the striped species from Lake Kivu (H. vittatus), and all striped Lake Victoria cichlids share the stripe haplotype. (B) ChromoPainter analysis visualizes the probability of haplotypic segments to be shared among lineages and thereby the ancestral relationships at the agrp2 locus of striped and nonstriped recipient species. Every horizontal line in the heatmap represents one haplotype of a recipient species. The structure of agrp2 is given at the bottom, and gray dashed lines connect an SNP in the ChromoPainter ancestry matrix to its relative region within agrp2. This heatmap visualization of the results illustrates a high probability of ancestry in darker colors. Candidate regions LV 1, LV 2 and the 538-bp region from (A) (region LVRS, indicated by dark orange dashed lines) of striped recipient species are more closely related to the riverine species (A. stappersii). In contrast, in nonstriped recipient species these regions are more closely related to the nonstriped donor species from Lake Kivu (H. gracilior). Candidate region LM (indicated at the top in blue) does not show a clear donor-recipient signal as there is also a high probability of relatedness of nonstriped Lake Victoria recipients to our control of a striped Lake Victoria donor species (H. sauvagei).
These analyses support that divergence between striped and nonstriped species in the radiations of East African cichlids is fueled by distinct cis-regulatory mechanisms controlling agrp2 expression, demonstrating that the recurrent involvement of the same gene does not necessarily mean that also the underlying causal mutations are the same.
The Causal Stripe Haplotype in Lake Victoria Evolved Prior to the Adaptive Radiation
The finding of a single haplotype associated with stripes across all Lake Victoria species in our data set is particularly interesting, as it suggests recruitment from ancestral standing genetic variation that was already present prior to the Lake Victoria cichlid radiation. To test this hypothesis, we analyzed the agrp2 locus in five species from ancestral lineages that are known (Verheyen et al. 2003) to have diverged before the onset of the adaptive radiation in Lake Victoria (i.e., from Lake Kivu). The more distantly related lineages include nonstriped species from Lake Kivu (“Haplochromis” gracilior) and Lake Edward (Thoracochromis pharyngalis). Two more closely related lineages include striped and nonstriped species endemic to Lake Kivu (H. vittatus and H. paucidens) and a striped riverine haplochromine species (Astatotilapia stappersii) from Kalambo River and Rusizi River (Greenwood 1979; Seehausen et al. 2003; Meier, Marques, et al. 2017), which form a connection between Lake Kivu and Lake Tanganyika. Horizontal stripes are only present in the more closely related lineages (fig. 1; Luc et al. 2001; McGee et al. 2016; Meier, Marques, et al. 2017). To test whether the causal alleles underlying stripe pattern divergence were already present in these more ancient haplochromine cichlid lineages, we analyzed the agrp2 locus in all striped ancestral species as well as in H. gracilior which was previously proposed as the source population of the Lake Victoria radiation (Verheyen et al. 2003).
First, we calculated FST between striped and nonstriped phenotypes of Lake Victoria cichlids, and the ancestral lineages. From the most differentiated region (region LVRS, 538 bp, FST = 0.88) we built a haplotype network. To reveal the evolutionary origin of the causal variants in the Lake Victoria superflock, we added species from Lake Malawi to the haplotype network (fig. 4A). The Lake Victoria superflock is a group of 700 haplochromine cichlid species endemic to the region around Lake Victoria and nearby western rift lakes in East Africa (Meyer et al. 1990; Verheyen et al. 2003; Seehausen 2006; Genner et al. 2007). The haplotype network shows that striped and nonstriped Lake Malawi cichlids have different haplotypes than striped Lake Victoria species (fig. 4A), as already suggested by the results above (fig. 2D and E). Yet, striped Lake Victoria species share the same haplotype with the two striped species of the ancestral lineages of the Lake Victoria radiation (riverine A. stappersii and H. vittatus from the older Lake Kivu). We can, therefore, conclude that the cis-regulatory interval (fig. 2A and B) must have evolved after their split from their common ancestor with Lake Malawi cichlids (2–4 Ma) but before their major radiation into the endemic species flocks of Lake Victoria and Lake Kivu (>0.5 Ma, the age of Lake Kivu; Verheyen et al. 2003). To identify the ancestral lineage from which the haplotype at the agrp2 locus originated, we used ChromoPainter (Lawson et al. 2012). For several Lake Victoria species, we calculated the per-site probability of ancestry along haplotypes (fig. 4B). Species from Lake Victoria acted as recipients with three ancestral striped and nonstriped species acting as donors (i.e., ancestral haplotypes that are sources of recipient haplotypes). In total, we used three striped and nonstriped recipient species each and ran two separate analyses for every striped and nonstriped recipient haplotype: Apart from the three ancestral lineages that acted as donors in the first analysis, all striped species had one nonstriped within-lake donor acting as a control, whereas all nonstriped species had one striped within-lake donor as control. Thereby, four donor species were competing in every analysis where a single recipient haplotype was tested—three ancestral donors and one within-lake control. If the cis-regulatory interval would have evolved within Lake Victoria, we would expect high per-site probability of ancestry for these within-lake comparisons—this is not the case. This result underlines the role of old standing genetic variation from ancestral lineages in driving the repeated evolution of stripes in the Lake Victoria cichlid species flock. We found strong evidence that the cis-regulatory interval in striped Lake Victoria species (candidate regions LV 1 and LV 2; fig. 2B) is most closely related to the riverine A. stappersii whereas other segments of the agrp2 locus are more closely related to the striped species from Lake Kivu (H. vittatus; fig. 4B).
The region of highest differentiation between striped and nonstriped species of the whole Lake Victoria superflock (fig. 4A) overlaps with candidate region LV 1. This region shows a higher probability of ancestry from the striped donor species of the riverine haplochromine (A. stappersii) than from all other striped donors. Pairwise comparisons between the striped and nonstriped species from Lake Victoria as well as its sister lineages revealed the same highly differentiated SNPs (fig. 3). Therefore, incomplete lineage sorting due to ancestral standing genetic variation that was introduced into the lake by the haplochromine founders is the most parsimonious explanation for the recurrent evolution of stripes in the Lake Victoria species flock.
Several recent studies across a wide range of study systems suggested that rapid speciation often involves “old genetic variants” upon which selection can act (Elmer and Meyer 2011; Machado-Schiaffino et al. 2017; Han et al. 2017; Meier, Marques, et al. 2017; Van Belleghem et al. 2017; Cameron and Whitfield 2019; Edelman et al. 2019; Jiggins 2019; Lewis et al. 2019; Marques et al. 2019; Kautt et al. forthcoming). By a comprehensive analysis of the “stripe locus” with its well-resolved genotype–phenotype connection, we provide additional insights into how ancestral standing genetic variation at the root of adaptive radiations can facilitate rapid phenotypic divergence within species flocks.
By tracing the evolutionary history of highly associated variants, our study sheds light on the origin of the genetic basis of horizontal stripes, an adaptive phenotype that evolved repeatedly within the hundreds of species of the East African cichlid radiations (Seehausen and Alphen 2001). Our findings show how different cis-regulatory regions of the same gene, agrp2, underlie rapid phenotypic divergence in the adaptive radiations of haplochromine cichlid fishes. We discovered that ancestral variants that form the genetic basis for stripe phenotypes in the Lake Victoria radiation predate the lake colonization and were introduced into it by the ancestors of this species flock and thereby allowed the repeated gain and loss of horizontal stripes within <100,000 years. In this radiation of more than 500 species, ancestral variants with an identified phenotypic effect (Kratochwil et al. 2018) permitted the repeated phenotypic diversification and explosive speciation that characterizes the Lake Victoria cichlid fish adaptive radiation.
Materials and Methods
Experimental Model and Sampling Details
This study was performed in accordance with the rules of the animal research facility (T-16/13) of the University of Konstanz and the animal protection authorities of the State of Baden-Württemberg.
We obtained whole-genome resequencing data from mostly wild-caught individuals from several different sources (see supplementary table S1, Supplementary Material online). The whole-genome samples from Malinsky et al. (2018), Meier, Sousa, et al. (2017), Meier, Marques, et al. (2017), and McGee et al. (2016) were obtained from wild-caught individuals. The genomes from Brawand et al. (2014) were inbred individuals from different laboratories.
Additionally, we sequenced 83 samples using target enrichment and 15 samples using whole-genome resequencing. These samples were obtained from wild-caught individuals from commercial breeders and maintained in the animal research facility of the University of Konstanz.
For most species, we sampled one individual per species to obtain a comprehensive data set that includes all major lineages. The reason why we have two samples of some species is that we used a combination of available genomes and target enrichment data for the agrp2 locus. This also allowed us to verify that different sequencing approaches did not introduce any biases during the downstream analyses (see supplementary table S1, Supplementary Material online).
Generally, we were very conservative with the phenotyping and classified a species as “striped” if either the male or the female possessed a horizontal stripe along the lateral side. All specimens that we sampled for this study were phenotyped using a photography chamber as described in Kratochwil et al. (2018). To the best of our knowledge, there are few polymorphic species and these exceptions (Neolamprologus buescheri, H. phythophagus) we sampled ourselves and documented the phenotype accordingly. In our analyses, these samples appear separately as if belonging to a striped and a nonstriped species.
Species Names and Assignment
Several of the analyzed species have different names across the literature. Because no commonly accepted taxonomy of cichlids is available, we added a column “Current Status” in supplementary table S1, Supplementary Material online, which is based on the current classification of the “Catalog of Fishes of the California Academy” (Fricke 2020).
Target Enrichment Data
Target enrichment data were produced using customized 120-nt baits with ∼3× flexible tiling density by MYBaits for the 270-kb interval around agrp2. Baits were designed from the P. nyererei reference genome (Brawand et al. 2014) which was curated by filling gaps of the genome assembly using Sanger sequencing reads so that the modified genome now contains the “stripe interval” (Kratochwil et al. 2019). This version of the P. nyererei genome is available in Dryad: https://doi.org/10.5061/dryad.bnzs7h467 (Kratochwil et al. 2020).
DNA was extracted either from muscle tissue or from fin clips stored in EtOH following the DNeasy Blood & Tissue Protocol (QIAGEN) or Genaxxon Genomic DNA Purification Mini Spin Column Kit (Genaxxon Bioscience GmbH), respectively. For library preparation, we used the Illumina TruSeq Nano HT Library Preparation Kit (Illumina Inc.) following the manufacturer’s guidelines. For the baits, we followed the MYBaits manual v3 (https://arborbiosci.com/wp-content/uploads/2017/10/MYbaits-manual-v3.pdf, last accessed September 25, 2020) and hybridized the probes at 65 °C for 22 h. Probes were sequenced in paired-end mode on a HiSeq 2500 system.
Whole-Genome Resequencing
DNA was extracted as explained above and DNA concentration was measured with fluorescence spectrophotometry by Qubit (Invitrogen). For library preparation, we used the Illumina TruSeq Nano HT Library Preparation Kit (Illumina Inc.) following the manufacturer’s guidelines. Samples were run on a Bioanalyser 12000 Chip to assure a high quality of DNA libraries and afterwards amplified using PCR. Finally, size distributions of all libraries were checked on a Bioanalyser HS chip before pooling them equimolarly. Sequencing was performed in paired-end mode (151PE) on a Hiseq X Ten platform (Illumina Inc.). Quality of the sequenced reads was assessed using MultiQC (Ewels et al. 2016).
The short-read data have been archived in the NCBI SRA database under the bioproject accession number PRJNA649899.
Quality Control and Statistical Analysis
Illumina adapters were trimmed from the raw fastq reads using picard v2.17.11 (http://broadinstitute.github.io/picard, last accessed September 25, 2020), and reads were mapped to the curated P. nyererei reference genome (Kratochwil et al. 2019, 2020) using bwa mem v0.7.12 (Li and Durbin 2009) and duplicate reads were marked with picard v2.17.11. Variants were called using the standard filter (--min-mapping-quality 30 --min-base-quality 20 --min-supporting-allele-qsum 0 --genotype-variant-threshold 0) and population options (population-based Bayesian inference model) in freebayes v1.1.0 (Garrison and Marth 2012). We decomposed multiple nucleotide polymorphisms in the VCF file into one SNP per line using a custom python script. The resulting VCF file was then hard-filtered using common hard filters from vcflib’s vcffilter (“QUAL > 1 & QUAL/AO > 10 & SAF > 0 & SAR > 0 & RPR > 1 & RPL > 1”), where “QUAL” refers to the quality of the variant site and thus removes really bad sites; “QUAL/AO > 10” requires an additional contribution of each alternative allele observation of 10 log units (∼Q10 per read); “SAF > 0 & SAR > 0” requires that alternative allele observations are present on both strands; and “RPR > 1 & RPL > 1” requires that at least two reads with alternative allele observations are placed toward each side of the variant site. Additionally, we used VCFtools v0.1.15 (Danecek et al. 2011) to remove indels, include only biallelic sites (--max-alleles 2) and exclude sites that are missing in more than 5% of the samples (--max-missing 0.05).
Finally, the filtered VCF file was normalized using vt normalize (Tan et al. 2015). Mean effective sequencing depth, estimated from filtered VCF files using samtools flagstat (Li et al. 2009) can be found in supplementary table S1, Supplementary Material online.
Next, we extracted phase-informative reads using quality filters --base-quality 13 and --read-quality 10 before phasing with SHAPEIT v2.r790 (Delaneau et al. 2012) and generated individual consensus fasta sequences with a custom python script. The consensus base was only kept when the site depth was above 5× coverage.
To calculate mean absolute genetic divergence (dXY) and mean relative genetic differentiation (FST) between striped and nonstriped species, we used the program Saguaro (Zamani et al. 2013). With no prior assumption about the relatedness of the species, Saguaro creates local distance matrices for each region of the genome. This method infers local relationships among individuals in the form of genetic distance matrices and assigns segments across the genomes to these topologies. Thereby, it is possible that a single SNP that is alternatively fixed/highly associated between two populations results in a candidate region for high relative genetic differentiation.
To identify regions within the stripe interval that differ between stripe phenotypes, we ran TWISST (Van Belleghem et al. 2017) for each lake separately. To reduce computation time, we used a subset of six species per lake (three striped and three nonstriped) resulting in 15 unrooted topologies. For this, we followed the authors’ recommendations (https://github.com/simonhmartin/twisst/, last accessed September 25, 2020), which involved variant calling with GATK’s Haplotype Caller (Poplin et al. 2018), filtering with VCFtools v0.1.15 (Danecek et al. 2011), and phasing with beagle 4 (Browning and Browning 2007). Finally, we constructed neighbor-joining trees for SNP windows (window size 50) in PhyML v3.1 (Guindon et al. 2010).
The curated P. nyererei reference genome assembly (Kratochwil et al. 2019) had a 26,936-bp zero coverage assembly gap on scaffold 3 that we removed from all plots as we also did not find this region in closely related species (Maylandia zebra, A. calliptera).
Next, we inferred a gene tree for the loci with the highest FST values (candidate regions LM, LV 1, and LV 2) using jModelTest v2.1.1 (Darriba et al. 2012) to find the appropriate substitution model and BEAST 2 (Bouckaert et al. 2014).
To compare the topology of the gene trees with a species tree, we inferred the phylogenetic relationships using 33 genomes (fig. 1). In brief, we mapped the 33 whole-genome sequences to the Oreochromis niloticus genome assembly (NCBI: GCA_000188235.1) which is more complete (higher scaffold N50) than the P. nyererei genome. Variant calling and filtering steps were performed as described earlier. In the python script used to generate individual consensus fasta sequences, we applied a maximum missingness filter of 0.75 that excludes sites on the basis of the proportion of missing data. Then, from each genome we extracted loci with a maximum physical extent of 3,000 bp each of which a minimum of 2,000 sites had to be covered. These genome-wide loci were selected randomly, requiring a minimum distance of 100 kb between loci resulting in a total of 6,545 genome-wide loci. Next, we inferred single-gene trees of all loci using IQ-tree 1.6.9 (Nguyen et al. 2015) with the ModelFinder option (Kalyaanamoorthy et al. 2017) for automatic selection of the appropriate model of evolution and with 100 rounds of ultrafast bootstrapping (Hoang et al. 2018) and estimation of the Shimodaira–Hasegawa-like approximate likelihood ratio test (Guindon et al. 2010), respectively. Ultimately, we built the species tree using all 6,545 gene trees in ASTRAL-III (Zhang et al. 2018). All trees were illustrated with FigTree v1.4.0.
To provide additional supporting evidence that the substitutions close to and within the 5′-UTR of agrp2 in Lake Malawi mbuna could have an effect on agrp2 transcription, we screened the two divergent haplotypes for TFBSs using JASPAR (Fornes et al. 2020). For this, we extracted the flanking sequences of each SNP (±10 bp) within candidate region LM (fig. 3 and supplementary fig. S2, Supplementary Material online) and screened for TFBSs above a conservative threshold of 0.85 (Kwon et al. 2012) of the relative matrix score in at least one of the two species. The difference between relative scores (delta Pdem − Pcya) of Ps. demasoni and Ps. cyaneorhabdos serves as an indicator of differential regulation of agrp2 between the nonstriped and striped species. Supplementary table S2, Supplementary Material online, gives a summary of all results as well as location of gene expression which we collected from ZFIN (https://zfin.org/, last accessed September 25, 2020).
We performed the same analysis for region LV 1 using the nonstriped species P. nyererei, Pnye, and striped H. sauvagei, Hsau. We focused on LV 1, as it had the strongest association, also when we included closely related riverine and Lake Kivu lineages (see overlapping region LVRS in fig. 4).
Since we find the association of noncoding regions within agrp2 to stripe phenotypes in all striped species from Lake Victoria in our data set, we traced the evolutionary origin of those haplotypes. First, we included striped and nonstriped ancestral lineages of Lake Victoria and repeated the analysis of mean relative genetic differentiation (FST) between striped and nonstriped species. The resulting region (labeled LVRS for Lake Victoria Region Superflock) of highest differentiation overlaps with the previous identified region LV1, however, is slightly shorter (538 bp instead of 687 bp). We used the R package pegas (Paradis 2010) to plot a haplotype network from this 538-bp region and included the Lake Malawi mbuna to show that the stripe haplotype is not shared between radiations.
Lastly, we used ChromoPainter (Lawson et al. 2012) to elucidate haplotype relationships within the agrp2 locus in Lake Victoria. ChromoPainter models each recipient haplotype as a mosaic of the donor haplotypes while capturing which donors are essential to explain the recipient. There, we used different striped and nonstriped donors (ancestral species which are the sources of admixture) and three striped and three nonstriped recipient species, which represent the recipients of admixture. For visualization, we used the pheatmap function (Kolde 2019) in R. Scripts are available in the GitHub repository (https://github.com/sabineurban, last accessed September 25, 2020).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was funded by fellowships of the International Max Planck Research School for Organismal Biology (to S.U.), the Swiss National Science Foundation (P300PA_177852 to A.N.), the Elite-Program-for-Postdocs, Baden-Württemberg Foundation, the Deutsche Forschungsgemeinschaft (DFG, KR 4670/2-1 and KR 4670/4-1 to C.F.K.), and the European Research Council (ERC Advanced Grant, GenAdap 293700 to A.M.). Computations were carried out on resources provided by the Scientific Compute Cluster of the University of Konstanz. The authors thank Jannik Beninde, Jan Gerwin, Yipeng Liang, Stefan Gerlach, and the staff of the animal research facility of the University of Konstanz for their valuable help.
Author Contributions
S.U. wrote the article with contributions from all authors; S.U. and C.F.K. conducted sample collection; S.U. did bench work; S.U. and A.N. conducted analyses; C.F.K. designed the study. C.F.K. and A.M. supervised the study.
Data Availability
Fastq raw reads of all samples sequenced for this study (supplementary table S1, Supplementary Material online) are deposited at the NCBI Sequence Read Archive under the bioproject accession number PRJNA649899.
References
- Albertson RC, Powder KE, Hu Y, Coyle KP, Roberts RB, Parsons KJ.. 2014. Genetic basis of continuous variation in the levels and modular inheritance of pigmentation in cichlid fishes. Mol Ecol. 23(21):5135–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, Burns SC, Penalva LO.. 2012. Before it gets started: regulating translation at the 5' UTR. Comp Funct Genomics. 2012:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett LW, Fletcher S, Wilton SD.. 2012. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci. 69(21):3613–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RD, Schluter D.. 2008. Adaptation from standing genetic variation. Trends Ecol Evol. 23(1):38–44. [DOI] [PubMed] [Google Scholar]
- Bassham S, Catchen J, Lescak E, von Hippel FA, Cresko WA.. 2018. Repeated selection of alternatively adapted haplotypes creates sweeping genomic remodeling in stickleback. Genetics 209:921–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513(7518):375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL.. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81(5):1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron SA, Whitfield JB.. 2019. Shift in temporal and spatial expression of Hox gene explains color mimicry in bees. Proc Natl Acad Sci U S A. 116:11573–11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceinos RM, Guillot R, Kelsh RN, Cerda-Reverter JM, Rotllant J.. 2015. Pigment patterns in adult fish result from superimposition of two largely independent pigmentation mechanisms. Pigment Cell Melanoma Res. 28(2):196–209. [DOI] [PubMed] [Google Scholar]
- Clabaut C, Salzburger W, Meyer A.. 2005. Comparative phylogenetic analyses of the adaptive radiation of Lake Tanganyika cichlid fish: nuclear sequences are less homoplasious but also less informative than mitochondrial DNA. J Mol Evol. 61(5):666–681. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Peichel CL, Nereng K, Blackman BK, Shapiro MD, Schluter D, Kingsley DM.. 2004. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2(5):E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. ; 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danley PD, Kocher TD.. 2001. Speciation in rapidly diverging systems: lessons from Lake Malawi. Mol Ecol. 10(5):1075–1086. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF.. 2012. A linear complexity phasing method for thousands of genomes. Nat Methods. 9(2):179–181. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Raible DW, Moon RT.. 2000. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 14:158–162. [PMC free article] [PubMed] [Google Scholar]
- Edelman NB, Frandsen PB, Miyagi M, Clavijo B, Davey J, Dikow RB, Garcia-Accinelli G, Van Belleghem SM, Patterson N, Neafsey DE, et al. 2019. Genomic architecture and introgression shape a butterfly radiation. Science 366(6465):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, , Meyer A. 2011. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends in Ecology & Evolution. 26(6):298–306. 10.1016/j.tree.2011.02.008 [DOI] [PubMed] [Google Scholar]
- Elmer KR, Fan S, Kusche H, Spreitzer ML, Kautt AF, Franchini P, Meyer A.. 2014. Parallel evolution of Nicaraguan crater lake cichlid fishes via non-parallel routes. Nat Commun. 5(1):5168. [DOI] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M.. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32(19):3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranasic D, et al. 2020. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48:D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke R, , Eschmeyer WN, , Van der Laan R. 2020. Eschmeyer’s Catalog of Fishes: genera, species, references. Available from: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp. Accessed January 3, 2020. San Francisco (CA). [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907.
- Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, Turner GF.. 2007. Age of cichlids: new dates for ancient lake fish radiations. Mol Biol Evol. 24(5):1269–1282. [DOI] [PubMed] [Google Scholar]
- Genner MJ, Turner GF.. 2005. The mbuna cichlids of Lake Malawi: a model for rapid speciation and adaptive radiation. Fish Fish. 6(1):1–34. [Google Scholar]
- Greenwood PH. 1979. Towards a phyletic classification of the ‘genus’ Haplochromis (Pisces, Cichlidae) and related taxa. Part I. Bull Brit Mus (Nat Hist). 35:265–322. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Han F, Lamichhaney S, Grant BR, Grant PR, Andersson L, Webster MT.. 2017. Gene flow, ancient polymorphism, and ecological adaptation shape the genomic landscape of divergence among Darwin's finches. Genome Res. 27(6):1004–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ML, Baxter LL, Loftus SK, Pavan WJ.. 2010. Sox proteins in melanocyte development and melanoma. Pigment Cell Melanoma Res. 23(4):496–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol. 22(18):4606–4618. [DOI] [PubMed] [Google Scholar]
- Henning F, Lee HJ, Franchini P, Meyer A.. 2014. Genetic mapping of horizontal stripes in Lake Victoria cichlid fishes: benefits and pitfalls of using RAD markers for dense linkage mapping. Mol Ecol. 23(21):5224–5240. [DOI] [PubMed] [Google Scholar]
- Henning F, Meyer A.. 2014. The evolutionary genomics of cichlid fishes: explosive speciation and adaptation in the postgenomic era. Annu Rev Genom Hum Genet. 15(1):417–441. [DOI] [PubMed] [Google Scholar]
- Hines HM, Counterman BA, Papa R, Albuquerque de Moura P, Cardoso MZ, Linares M, Mallet J, Reed RD, Jiggins CD, Kronforst MR, et al. 2011. Wing patterning gene redefines the mimetic history of Heliconius butterflies. Proc Natl Acad Sci U S A. 108(49):19666–19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS.. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irisarri I, Singh P, Koblmuller S, Torres-Dowdall J, Henning F, Franchini P, Fischer C, Lemmon AR, Lemmon EM, Thallinger GG, et al. 2018. Phylogenomics uncovers early hybridization and adaptive loci shaping the radiation of Lake Tanganyika cichlid fishes. Nat Commun. 9(1):3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins CD. 2019. Can genomics shed light on the origin of species? PLoS Biol. 17(8):e3000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TC, Kelts K, Odada E.. 2000. The holocene history of Lake Victoria. AMBIO 29(1):2–11. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautt AF, Kratochwil CF, Nater A, Machado-Schiaffino G, Olave M, Henning F, Torres-Dowdall J, Härer A, Hulsey CDFranchini P, et al. Forthcoming. Contrasting signatures of genomic divergence during sympatric speciation. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblmüller S, Sefc KM, Sturmbauer C.. 2008. The Lake Tanganyika cichlid species assemblage: recent advances in molecular phylogenetics. Hydrobiologia 615(1):5–20. [Google Scholar]
- Kocher TD. 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 5(4):288–298. [DOI] [PubMed] [Google Scholar]
- Kolde R. 2019. Pheatmap: Pretty Heatmaps. Version 1.0.12. Available from: https://cran.r-project.org/web/packages/pheatmap/index.html. Accessed September 25, 2020.
- Kratochwil CF, Liang Y, Gerwin J, Woltering JM, Urban S, Henning F, Machado-Schiaffino G, Hulsey CD, Meyer A.. 2018. Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362(6413):457–460. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Liang Y, Urban S, Torres-Dowdall J, Meyer A.. 2019. Evolutionary dynamics of structural variation at a key locus for color pattern diversification in cichlid fishes. Genome Biol Evol. 11(12):3452–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil CF, Liang Y, Urban S, Torres-Dowdall J, Meyer A.. 2020. Evolutionary dynamics of structural variation at a key locus for color pattern diversification in cichlid fishes. v4. Dryad, Dataset: 10.5061/dryad.bnzs7h467. [DOI] [PMC free article] [PubMed]
- Kratochwil CF, Meyer A.. 2015. Closing the genotype-phenotype gap: emerging technologies for evolutionary genetics in ecological model vertebrate systems. Bioessays 37(2):213–226. [DOI] [PubMed] [Google Scholar]
- Kuraku S, Meyer A.. 2008. Genomic analysis of cichlid fish ‘natural mutants’. Curr Opin Genet Dev. 18(6):551–558. [DOI] [PubMed] [Google Scholar]
- Kwon AT, , Arenillas DJ, , Hunt RW, , Wasserman WW. 2012. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3. 2(9):987–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhaney S, Han F, Berglund J, Wang C, Almen MS, Webster MT, Grant BR, Grant PR, Andersson L.. 2016. A beak size locus in Darwin's finches facilitated character displacement during a drought. Science 352(6284):470–474. [DOI] [PubMed] [Google Scholar]
- Lavallee-Adam M, Cloutier P, Coulombe B, Blanchette M.. 2017. Functional 5' UTR motif discovery with LESMoN: local Enrichment of Sequence Motifs in biological Networks. Nucleic Acids Res. 45(18):10415–10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DJ, Hellenthal G, Myers S, Falush D.. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8(1):e1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Kalcheim C.. 1999. The neural crest. 2nd ed.Cambridge: Cambridge University Press. [Google Scholar]
- Lewis JJ, Geltman RC, Pollak PC, Rondem KE, Van Belleghem SM, Hubisz MJ, Munn PR, Zhang L, Benson C, Mazo-Vargas A, et al. 2019. Parallel evolution of ancient, pleiotropic enhancers underlies butterfly wing pattern mimicry. Proc Natl Acad Sci U S A. 116(48):24174–24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luc DV, Jos S, Thys van den Audenaerde D.. 2001. An annotated checklist of the fishes of Rwanda (East Central Africa), with historical data on introductions of commercially important species. J East Afr Nat Hist. 90:41–68. [Google Scholar]
- Machado-Schiaffino G, , Kautt AF, , Torres-Dowdall J, , Baumgarten L, , Henning F, , Meyer A. 2017. Incipient speciation driven by hypertrophied lips in Midas cichlid fishes?. Mol Ecol. 26(8):2348–2362. [DOI] [PubMed] [Google Scholar]
- Malinsky M, Svardal H, Tyers AM, Miska EA, Genner MJ, Turner GF, Durbin R.. 2018. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat Ecol Evol. 2(12):1940–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manceau M, Domingues VS, Mallarino R, Hoekstra HE.. 2011. The developmental role of Agouti in color pattern evolution. Science 331(6020):1062–1065. [DOI] [PubMed] [Google Scholar]
- Marques DA, Meier JI, Seehausen O.. 2019. A combinatorial view on speciation and adaptive radiation. Trends Ecol Evol. 34(6):531–544. [DOI] [PubMed] [Google Scholar]
- McGee MD, Neches RY, Seehausen O.. 2016. Evaluating genomic divergence and parallelism in replicate ecomorphs from young and old cichlid adaptive radiations. Mol Ecol. 25(1):260–268. [DOI] [PubMed] [Google Scholar]
- Meier JI, Marques DA, Mwaiko S, Wagner CE, Excoffier L, Seehausen O.. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat Commun. 8(1):14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier JI, Sousa VC, Marques DA, Selz OM, Wagner CE, Excoffier L, Seehausen O.. 2017. Demographic modelling with whole-genome data reveals parallel origin of similar Pundamilia cichlid species after hybridization. Mol Ecol. 26(1):123–141. [DOI] [PubMed] [Google Scholar]
- Meyer A. 1993. Phylogenetic relationships and evolutionary processes in East African cichlid fishes. Trends Ecol Evol. 8(8):279–284. [DOI] [PubMed] [Google Scholar]
- Meyer A, Kocher TD, Basasibwaki P, Wilson AC.. 1990. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature 347(6293):550–553. [DOI] [PubMed] [Google Scholar]
- Nelson TC, Cresko WA.. 2018. Ancient genomic variation underlies repeated ecological adaptation in young stickleback populations. Evol Lett. 2(1):9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M. 1991. Lake Tanganyika as an evolutionary reservoir of old lineages of East African cichlid fishes: inferences from allozyme data. Experientia 47(9):974–979. [Google Scholar]
- Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 23(1):263–286. [Google Scholar]
- Paradis E. 2010. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26(3):419–420. [DOI] [PubMed] [Google Scholar]
- Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. 2018. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv:201178.
- Protas ME, Patel NH.. 2008. Evolution of coloration patterns. Annu Rev Cell Dev Biol. 24(1):425–446. [DOI] [PubMed] [Google Scholar]
- Salzburger W. 2018. Understanding explosive diversification through cichlid fish genomics. Nat Rev Genet. 19(11):705–717. [DOI] [PubMed] [Google Scholar]
- Salzburger W, Mack T, Verheyen E, Meyer A.. 2005. Out of Tanganyika: genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol Biol. 5(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Meyer A.. 2004. The species flocks of East African cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften. 91(6):277–290. [DOI] [PubMed] [Google Scholar]
- Salzburger W, Meyer A, Baric S, Verheyen E, Sturmbauer C.. 2002. Phylogeny of the Lake Tanganyika cichlid species flock and its relationship to the Central and East African haplochromine cichlid fish faunas. Syst Biol. 51(1):113–135. [DOI] [PubMed] [Google Scholar]
- Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, Sanchez-Garcia I.. 2002. SLUG (SNAI2) deletions in patients with Waardenburg disease. Hum Mol Genet. 11(25):3231–3236. [DOI] [PubMed] [Google Scholar]
- Seehausen M, Alphen JJMV.. 2001. Evolution of colour patterns in East African cichlid fish. J Evol Biol. 12:514–534. [Google Scholar]
- Seehausen O. 2004. Hybridization and adaptive radiation. Trends Ecol Evol. 19(4):198–207. [DOI] [PubMed] [Google Scholar]
- Seehausen O. 2006. African cichlid fish: a model system in adaptive radiation research. Proc R Soc B. 273(1597):1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. 2015. Process and pattern in cichlid radiations–inferences for understanding unusually high rates of evolutionary diversification. New Phytol. 207(2):304–312. [DOI] [PubMed] [Google Scholar]
- Seehausen O, Koetsier E, Schneider MV, Chapman LJ, Chapman CA, Knight ME, Turner GF, van Alphen JJ, Bills R.. 2003. Nuclear markers reveal unexpected genetic variation and a Congolese-Nilotic origin of the Lake Victoria cichlid species flock. Proc R Soc Lond B. 270(1511):129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. 2013. The genetic causes of convergent evolution. Nat Rev Genet. 14(11):751–764. [DOI] [PubMed] [Google Scholar]
- Stiassny MLJ, Meyer A.. 1999. Cichlids of the rift lakes. Sci Am. 280(2):64–69. [Google Scholar]
- Sturmbauer C, Meyer A.. 1992. Genetic divergence, speciation and morphological stasis in a lineage of African cichlid fishes. Nature 358(6387):578–581. [DOI] [PubMed] [Google Scholar]
- Svardal H, Quah FX, Malinsky M, Ngatunga BP, Miska EA, Salzburger W, Genner MJ, Turner GF, Durbin R.. 2020. Ancestral hybridization facilitated species diversification in the Lake Malawi cichlid fish adaptive radiation. Mol Biol Evol. 37(4):1100–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Koblmüller S.. 2011. The adaptive radiation of cichlid fish in Lake Tanganyika: a morphological perspective. Int J Evol Biol. 2011:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A, Abecasis GR, Kang HM.. 2015. Unified representation of genetic variants. Bioinformatics 31(13):2202–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GF. 2007. Adaptive radiation of cichlid fish. Curr Biol. 17(19):R827–R831. [DOI] [PubMed] [Google Scholar]
- Turner GF, Seehausen O, Knight ME, Allender CJ, Robinson RL.. 2008. How many species of cichlid fishes are there in African lakes? Mol Ecol. 10(3):793–806. [DOI] [PubMed] [Google Scholar]
- Van Belleghem SM, Rastas P, Papanicolaou A, Martin SH, Arias CF, Supple MA, Hanly JJ, Mallet J, Lewis JJ, Hines HM, et al. 2017. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 1(3):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Otterloo E, Li W, Bonde G, Day KM, Hsu MY, Cornell RA.. 2010. Differentiation of zebrafish melanophores depends on transcription factors AP2 alpha and AP2 epsilon. PLoS Genet. 6(9):e1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen E, Salzburger W, Snoeks J, Meyer A.. 2003. Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science 300(5617):325–329. [DOI] [PubMed] [Google Scholar]
- Wagner CE, Keller I, Wittwer S, Selz OM, Mwaiko S, Greuter L, Sivasundar A, Seehausen O.. 2013. Genome-wide RAD sequence data provide unprecedented resolution of species boundaries and relationships in the Lake Victoria cichlid adaptive radiation. Mol Ecol. 22(3):787–798. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhao H, Yang J, Ma Y, Liu Z, Li C, Wang T, Yan Z, Du N.. 2019. MiR-150-5p regulates melanoma proliferation, invasion and metastasis via SIX1-mediated Warburg Effect. Biochem Biophys Res Commun. 515(1):85–91. [DOI] [PubMed] [Google Scholar]
- York RA, Patil C, Abdilleh K, Johnson ZV, Conte MA, Genner MJ, McGrath PT, Fraser HB, Fernald RD, Streelman JT.. 2018. Behavior-dependent cis regulation reveals genes and pathways associated with bower building in cichlid fishes. Proc Natl Acad Sci U S A. 115(47):E11081–E11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamani N, Russell P, Lantz H, Hoeppner MP, Meadows JR, Vijay N, Mauceli E, di Palma F, Lindblad-Toh K, Jern P, et al. 2013. Unsupervised genome-wide recognition of local relationship patterns. BMC Genomics 14(1):347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S.. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19(Suppl 6):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Song Y, Thompson DA, Madonna MA, Millhauser GL, Toro S, Varga Z, Westerfield M, Gamse J, Chen W, et al. 2010. Pineal-specific agouti protein regulates teleost background adaptation. Proc Natl Acad Sci U S A. 107(47):20164–20171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fastq raw reads of all samples sequenced for this study (supplementary table S1, Supplementary Material online) are deposited at the NCBI Sequence Read Archive under the bioproject accession number PRJNA649899.