Abstract
Traditional epitranscriptomics relies on capturing a single RNA modification by antibody or chemical treatment, combined with short-read sequencing to identify its transcriptomic location. This approach is labor-intensive and may introduce experimental artifacts. Direct sequencing of native RNA using Oxford Nanopore Technologies (ONT) can allow for directly detecting the RNA base modifications, although these modifications might appear as sequencing errors. The percent Error of Specific Bases (%ESB) was higher for native RNA than unmodified RNA, which enabled the detection of ribonucleotide modification sites. Based on the %ESB differences, we developed a bioinformatic tool, epitranscriptional landscape inferring from glitches of ONT signals (ELIGOS), that is based on various types of synthetic modified RNA and applied to rRNA and mRNA. ELIGOS is able to accurately predict known classes of RNA methylation sites (AUC > 0.93) in rRNAs from Escherichiacoli, yeast, and human cells, using either unmodified in vitro transcription RNA or a background error model, which mimics the systematic error of direct RNA sequencing as the reference. The well-known DRACH/RRACH motif was localized and identified, consistent with previous studies, using differential analysis of ELIGOS to study the impact of RNA m6A methyltransferase by comparing wild type and knockouts in yeast and mouse cells. Lastly, the DRACH motif could also be identified in the mRNA of three human cell lines. The mRNA modification identified by ELIGOS is at the level of individual base resolution. In summary, we have developed a bioinformatic software package to uncover native RNA modifications.
INTRODUCTION
The transcriptome is the collection of all RNA molecules present in a given cell that can be determined by high-throughput techniques, such as microarray analysis or RNA sequencing (RNA-seq) methods (1). Using next-generation sequencing (NGS) techniques, RNA-seq has been replacing microarray analysis, because the former can detect novel or unknown transcripts. Further, NGS enables transcriptome analysis with a higher dynamic range of expression levels than microarrays (2). With improved sample preparation methods and reduced sequencing costs, RNA-seq by NGS has become the method of choice to analyze transcriptomes.
The length of individual sequence reads generated with most NGS platforms ranges from 35 nucleotides (nt) to about 500 nt, so that single reads rarely cover a complete transcript, which, on average, is approximately a thousand nucleotides in bacteria, and can be much longer in eukaryotes. Accurate alignment and assembly of such short sequences depend on the availability of a reference genome; the identification of spliced isoforms, edited messenger RNA (mRNA), or gene-fusion transcripts remains a challenge (3). Further, methods using reverse transcription (RT) of RNA and polymerase chain reaction (PCR) amplification may introduce biases and artifacts (4). These shortcomings can be overcome by directly sequencing native RNA molecules using the Oxford Nanopore Technologies (ONT) platform (Oxford, UK). Direct RNA sequencing (dRNA-seq) without amplification can generate long reads, typically covering the full length of a transcript (5). The method can accurately quantify transcripts to analyze differential gene expression with a dynamic range comparable to traditional RNA-seq derived from short-read sequencing, while it enables accurate identification of the structure and boundaries of transcripts, including spliced products and polyadenylation (polyA) length (6,7).
An additional advantage of dRNA-seq is the detection of transcriptional modifications inferred from the current signal as the RNA molecule passes a nanopore: modified RNA molecules cause a characteristic temporary current blockade, enabling simultaneous detection of diverse modifications such as 5-methylcytosine (m5C) or N6-methyladenine (m6A) (5,7–9). Currently, there are >170 different types of RNA modifications that have been described within the prokaryote and eukaryote kingdoms and are collected in various databases (10–12). High-throughput sequencing coupled with methods to specifically enrich RNA modification products makes it possible to study the epigenetics of RNA, or its ‘epitranscriptome’ (13,14). However, current methods are labor-intensive and may introduce experimental artifacts or biased results and are accompanied with a relatively high false-positive rate (15). Moreover, the transcriptome-wide approach can identify only a few of the over 170 known types of RNA modifications, as they depend on specific antibodies or chemical treatments (16). Alternatively, many RNA modification can be quantified simultaneously and accurately by liquid chromatography–tandem mass spectrometry (LC–MS/MS) at the nucleotide level, but this unfortunately forfeits the position of the modification that is essential for comprehensibly describing the epitranscriptome (16).
Translation of the obtained electrical current signals into specific bases currently relies on either trained hidden Markov models (HMMs) or artificial neural network models that produce an accuracy of individual DNA reads of ∼90%, on average (17). We typically experience a read accuracy of ∼88% in RNA (6). The most commonly encountered errors are related to some type of base modification, presence of homopolymers, nucleic acid damage, or structural features of the nucleic acid molecules. Therefore, dissection and analysis of sequencing errors can potentially uncover RNA modifications and other structural information from native RNA sequences.
To more accurately capture this information, we developed the epitranscriptional landscape inferring from glitches of ONT signals (ELIGOS) software tool that predicts the presence of modified bases from a comparison of background error data derived from in vitro transcription (IVT) and RT sequence data. The output of the tool was verified with synthetic IVT by incorporation of modified bases commonly found in mRNA and with rRNA sequences from Saccharomyces cerevisiae, E. coli, and cells from a human cell line. After this verification, we demonstrate the use of ELIGOS to investigate the epitranscriptional landscape in yeast, mouse, and human cells.
MATERIALS AND METHODS
Sequencing of DNA and RNA templates on an ONT platform
All sequencing of DNA and RNA templates described here was performed on MinION Mk1B flow cells (ONT). For direct complementary DNA (dcDNA) sequencing, a library was produced from mRNA and polyA-tailed rRNA (described below) using the SQK-DCS108 kit (ONT), which includes an RT step, but no amplification step, to give double-strand DNA (dsDNA), after which the adaptor containing the motor protein was attached by ligation. The library was loaded directly onto a flow cell for sequencing. Preparation of the library for dRNA-seq was done with the SQK-RNA002 kit (ONT) and only required an RNA stabilization step by the formation of DNA–RNA hybrids through RT. After this, the motor protein was attached specifically to the RNA strands. Each library was loaded onto a flow cell for a sequencing run lasting 48 h. Both of the direct sequencing runs were performed on a single R9.4.1/FLO-MIN106 (ONT) flow cell.
The raw data generated by MinKNOW software (version 1.7.14; ONT) were converted from .fast5 files to base-called .fastq files using the local-based caller Guppy version 2.3.4 software (ONT). Only reads greater than 200 bases were considered for further analysis. The reads were aligned to reference sequences using Minimap2 version 2.17 software (18) to generate a BAM file. Each BAM file, together with reference sequences and transcript annotation files in BED12 format, was used to retrieve substitutions, insertions, and deletions of individual positions through the pysam module version 0.13 (https://github.com/pysam-developers/pysam).
Comparative error analysis and development of ELIGOS software
The ELIGOS software compares the error profile between native RNA sequences obtained with dRNA-seq and a reference, which can be IVT RNA, cDNA sequences, or the RNA background error model (rBEM). Moreover, ELIGOS can be used to compare native RNA sequences of different conditions directly to identify differential epitranscriptomes.
First, rBEMs were constructed to capture the systematic noise of dRNA-seq using nonmodified IVT sequences (human and synthetic, as described below). The Error of Specific Bases (ESB) count is defined as the frequency of the sum of substitutions, insertions, and deletions of individual positions, over the total mapped reads obtained from read alignment results based on the reference sequence. These were obtained for all possible sequence lengths (i.e. kmers) of 5 nt (1,024 pentamer bins, corresponding to the number of ribonucleotides that dwell in the nanopore during sequencing (19)) and calculated over the reference sequence for an individual 5mer bin by a sliding window of one ribonucleotide. We aggregated the ESB information at individual positions (l, left position; c, center or middle position; r, right position), where l1 = −1 from c, l2 = −2 from c, r1 = +1 from c, and r2 = +2 from c for all pentamer bins as long as they included at least 50 mapped reads, to produce a dataset called rBEM_k5. A different error profile was observed that was characteristic for reads derived from homopolymeric regions when compared with rBEM_k5. We then extended the rBEM_k5 on the 3′ end with two more nucleotides, r3 = +3 from c and r4 = +4 from c, to produce dataset rBEM_k5+2.
The difference of the percent ESB (%ESB) between native RNA and corresponding dcDNA, nonmodified IVT reference sequences, or rBEMs were evaluated using Fisher's exact test for a single 2 × 2 contingency table of independence to produce odds ratios and P-values. The statistical P-values were further adjusted for multiple testing using the Benjamini–Hogberg method. To capture the signal alteration characteristics that can be present on one ribonucleotide and/or its neighboring position, we performed statistical tests for three scenarios: (i) data for position c only were compared; (ii) data were extended with one nucleotide on both sides of c by aggregated ESB of positions l1, c, and r1 and (iii) data were extended with two nucleotides on both sides by aggregated ESB of positions l2, l1, c, r1 and r2. To capture the variations of error pattern within a 5mer at a specific position, the maximum value of odds ratio derived from the three scenarios will be reported as a recommended result. The statistical tests were performed by R suite software through the rpy2 python module. ELIGOS is written in Python 3 and is available at https://gitlab.com/piroonj/eligos2 and https://hub.docker.com/repository/docker/piroonj/eligos2.
The software was applied to the synthetic modified IVT RNA, rRNA, and mRNA sequences obtained with the materials described in the next section. For rRNA investigations, the .fastq files were aligned onto a reference genome sequence (for S. cerevisiae, genes NR_132209.1, NR_132215.1, NR_132213.1, and NR_132211.1 were combined; for E. coli, positions 232785–23568, 1046691–1048228 and 232576–232686 from NZ_KK583188.1 were combined; and for H. sapiens, genes NR_023363.1, NR_003287.4, NR_146119.1 and NR_145819.1 were combined) using minimap2 software version 2.17 (18) to obtain BAM files of the sequences.
For mRNA investigations, we performed an analysis of two published data sets of the human cell line CEPH1463 (7) and yeast (20) with our generated experimental datasets of mouse embryonic stem cells (mESCs) and human lung cells (H460, small airway epithelial cells [SAEC]) as described below. The analysis was performed using the reference genomes S288c for yeast, mm9 for mouse, and hg38 for human. All data generated in this study were deposited in the Sequence Read Archives (SRA) database (accession number SRP166020). Some interesting regions were explored at the signal-level through the re-squiggle signal approach using Tombo software version 1.5 (ONT; https://github.com/nanoporetech/tombo.git).
De novo motif discovery
The sequences of six bases surrounding the considered differential %ESB identified by ELIGOS were extracted based on the reference sequence and were analyzed using BaMM software (21) to identify conserved motifs and scan the locations of the identified motif using default parameters.
Genomic location of positions and transcripts comparison
The relative location of the considered positions with reference to the gene location was compared using bedtools version 2.25 (22) and the GenomicRanges package (23). The results were summarized in Venn diagrams using ChIPpeakAnno (24) or upset plots using UpsetR (25).
Standardized coordinate plot of identified motifs
We used Guitar package (26) for standardized coordinate plots to evaluate the key landmark of the identified motifs on the structure of the transcript. The bed file of the identified position of the individual motif and dRNA-seq alignment was used as the input to calculate the density of the population along with the structure of the transcript. The density of the motif was then normalized with dRNA-seq alignment density. The normalized density was plotted along with standardized structure of the transcript in R suite software.
Statistical analysis
The performance of ELIGOS prediction based on synthetic IVTs and rRNAs was evaluated by Receiver Operating Characteristic (ROC) curve analysis of odd ratios using plotROC R package (27). Wilcoxon signed-rank sum tests were used to test the difference of means between two considered populations. All statistical analysis was performed in R suite software.
IVTs used for sequencing
IVT luciferase gene with 5-methoxyuridine incorporated
The transcript of the luciferase gene, containing standard ribonucleotides with and without the incorporation of 5-methoxyuridine (5moU), was obtained using CleanCap Firefly Luciferase mRNA (TriLink Biotechnologies, San Diego, CA, USA). The IVT mRNA containing a poly-A tail was purified using AMPureXP beads (Beckman Coulter, Brea, CA, USA) and eluted using nuclease-free water.
Synthetic DNA templates and IVT with the incorporation of modified nucleotides
For a systematic analysis, we constructed synthetic double-stranded DNA templates through gBlocks Gene Fragments (Integrated DNA Technologies [IDT], Coralville, IA, USA) that were targeted to contain a particular modified base after IVT within a defined pentamer, as five ribonucleotides dwell in the nanopore during transit (19). All possible pentamers (45 or 1024 sequences) of the individual bases A, T, C and G were investigated. Pentamers targeted to contain a modified A in their IVT were flanked by two pentamers that contained no A but were otherwise designed randomly with restriction of sequence complexity (e.g. BBBBB, or B5) under gBlocks Gene Fragment criteria. Thus, for a template targeted to contain one to five modified A nucleotides in their transcript, 15mers were designed as B5(NNNNN containing at least one A)B5. Likewise, pentamers designed to contain modified U were flanked by V5, those with modified G by H5, and those with modified C were flanked by D5. The 1024 constructs contained a standard T7 promoter sequence (5′-TAATACGACTCACTATAG-3′) in the sense strand. Details of the template sequences are provided in Supplementary Information.
IVT was performed with these templates using the AmpliScribe T7-Flash Transcription Kit (Lucigen Middleton, WI, USA). For the production of transcripts containing modified bases, the individual nucleotide triphosphate was completely replaced by a modified version, for which m6A, N1-methyladenine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytidine (hm5C), 5-formylcytidine (f5C), and pseudouridine (psU) were used (TriLink Biotechnologies, San Diego, CA, USA), as well as 7-methylguanosine (m7G; Sigma-Aldrich, St. Louis, MO, USA) and inosine (Ino; IBA Lifesciences, Göttingen, Germany). Following the IVT reaction, the RNA was purified with RNeasy Mini kit (Qiagen, Germantown, MD, USA). A poly(A) tail was added using E. coli Poly(A) (New England Biolabs, Ipswich, MA, USA) following a published protocol (28) and then used for library preparation.
IVT of human mRNA
Approximately 5 μg of purified total RNA from a human papillary thyroid cancer cell line, KTC-1, was depleted for rRNAs using QIAseq FastSelect RNA Removal Kit (Qiagen, Germantown, MD, USA). To produce sense IVT RNA from mRNA, we followed the terminal continuation method developed by Che et al. (29). The depleted RNA was mixed with a polyT (20mers) primer and a primer containing a strong T7 promoter (30) (5′-GCC GGG AAT TTA ATA CGA CTC ACT ATA GCG CTG TTG GTG TGC T rGrGrG-3′). cDNA was generated using RT, and terminal continuation was performed with Maxima Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). The RNA was digested using RNAase Cocktail Enzyme Mix (Thermo Fisher Scientific), after which double-strand DNA (dsDNA) synthesis was performed using Long Amp Taq Master Mix (New England Biolabs). This dsDNA was used as the template for IVT, performed as described above with canonical (nonmodified) nucleoside triphosphate and the resulting IVT RNA was purified as above.
Culture condition and RNA extraction for direct rRNA sequencing
RNA was extracted from yeast cells, E. coli cells, and from the human KTC-1 cells to directly sequence the rRNAs. For yeast, the S. cerevisiae strain S288C was grown overnight at 30°C in 15 ml yeast extract-peptone-dextrose (YPD) medium containing 10 g/l yeast extract, 20 g/l peptone, and 20 g/l glucose. RNA was extracted using the ZymoBIOMICS Quick-RNA Fungal/Bacterial kit (Zymo Research, Irvine, CA, USA). For E. coli strain ATCC 11775 was cultured overnight at 37°C in 25 ml of Luria broth (LB), and following centrifugation, the cell pellet was resuspended in 250 μl water, to which 750 μl of TRIzol Reagent (Life Technologies, Carlsbad, CA, USA) was added. Following incubation for 5 min at room temperature, 200 μl of chloroform was added. The liquid phases were mixed by inverting the tube 15 times and then incubated for 10 min. Following centrifugation at 12,000 × g for 5 minutes at 4°C, 400 μl of the aqueous phase was removed, and the RNA it contained was cleaned using the Direct-zol kit (Zymo Research).
For the human cell line, KTC-1 was grown to 85×90% confluence in 10 cm dishes in Roswell Park Memorial Institute (RPMI) media supplemented with 10% fetal bovine serum (FBS; R & D Systems, Minneapolis, MN, USA) using standard techniques. RNA isolation was performed with the Direct-zol RNA mini prep Kit (Zymo Research). Total RNA was eluted in 20 μl RNase/DNase free water and stored at –80°C. A poly(A) tail was added as described above. As most RNA in these samples represented rRNA, the template was completely sequenced to obtain rRNA reads.
Culture conditions and RNA extraction for direct mRNA sequencing
Yeast heat shock
S. cerevisiae strain S288C was grown on YPD (10 g/l yeast extract, 20 g/l peptone, 10 g/l glucose) for 12 h at 30°C. One aliquot of the cultured yeast cells was subjected to heat shock (45°C for 1 h) while the reference was kept at 30°C for an hour as a control. After the treatment, the cells were collected and immediately processed. RNA was extracted using the RNeasy Mini kit.
Mouse cells
Mettl3 knockout and control mESCs were provided by Dr Howard Y. Chang (Stanford University, Stanford, CA) (31). Mettl14 knockout and wildtype mESCs were provided by Dr Yawei Gao (Tongji University, Shanghai, China). Cells were maintained in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 15% FBS, 1% nucleosides (100×), 1 mM l-glutamine, 1% nonessential amino acids, 0.1 mM 2-mercaptoethanol, 1000 U/ml Leukemia Inhibitory Factor (LIF), 3 μM CHIR99021 and 1 μM PD0325901 in 37°C and 5% CO2. RNA was extracted on the collected cells using the RNeasy Mini kit.
Lung cell lines and culture
Human lung cancer cells H460 (ATCC, HTB-177) were cultured in RPMI 1640 medium (Corning Inc., Corning, NY, USA) supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (Corning) and subcultured twice per week (32). Human SAEC (CC-2547; Lonza Group, Basel, Switzerland) were cultured in SAGM Small Airway Epithelial Cell Growth Medium BulletKit (Lonza Group) and subcultured every 5–7 days following the manufacture's guidelines; the medium was refreshed with pre-equilibrated medium every 2 days, and these cells underwent up to five subcultures. Both cell lines were maintained as monolayer culture at 37°C and 5% CO2 in a humidified incubator. The cells were grown to ∼70% confluence and then dislodged using Trypsin/EDTA (Corning) and Trypsin Neutralizing Solution (Thermo Fisher Scientific), centrifuged and washed with phosphate buffer saline, and cell pellets were collected. RNA was extracted from the collected cells using RNeasy Mini kit.
RESULTS
Distinguishing modified RNA bases from sequencing errors
The nanopore sequencing signal can be affected by the 3D structures of an RNA template and by the presence of modified ribonucleotides; both of these can lead to systematic, position-specific sequencing errors, in addition to other stochastic errors in base calling. Because most of modified bases are absent when RNA is converted into cDNA, we anticipated that an in-depth analysis of sequencing errors in RNA and its corresponding modification-free cDNA might be allowed to differentiate between the presence of modified bases and stochastic errors. In a pilot experiment, we mimicked posttranscriptional modifications of RNA by sequencing IVT of a luciferase gene that had been produced with 5moU. Sequencing signals obtained with this modified mRNA (dRNAO) were compared to that of the corresponding cDNA by direct sequencing (dcDNAO) and to RNA produced with unmodified uridine by direct sequencing (dRNAU).
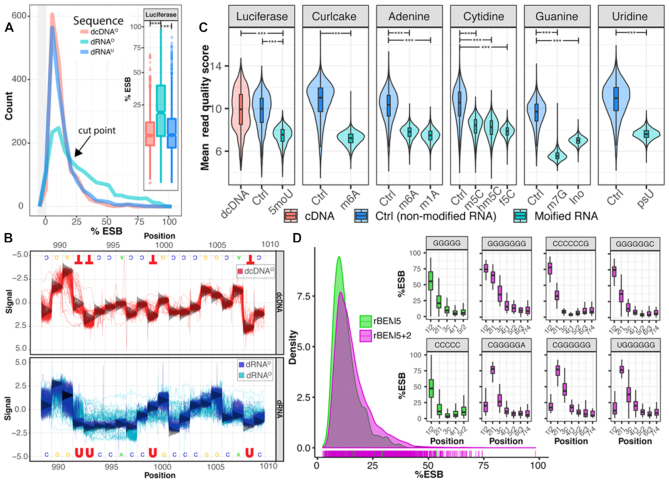
Figure 1A shows that incorporation of 5moU into the RNA resulted in reads with significantly higher %ESB than dcDNAO (P < e−60) or dRNAU (P < 1e−100). Notably, for values up to approximately 25%, the distributions of %ESB for both dRNAU and dcDNAO were overlapping and higher than those for dRNAO, but for values above 25%, dRNAO reported significantly higher %ESB (Figure 1A).
Figure 1.
Characteristics of direct sequencing of IVT RNA. (A) The distribution of the %ESB of luciferase IVT containing 5moU (dRNAO) differs significantly from that of cDNA derived from this (dcDNAO) and from direct sequencing of unmodified transcript (dRNAU), with ** P < e−60, ***P < e−100 (insert). The black arrow indicates at which frequency of %ESB higher values are found in dRNAO than in the other two templates. The gray area to the left of the plot represents the histogram of the first bin around zero. (B) Re-squiggled signal plots of a selected region of the luciferase gene obtained with dcDNAO template (top), and overlaid signals obtained with dRNAU (blue) and dRNAO (cyan) (bottom). The vertical, bell-shaped curves at each base position represent the distribution of the standard canonical model signals of base calling for either template. (C) Violin-boxplots of mean read quality scores of the IVT RNAs derived from synthetical constructs of DNA templates from which RNAs were produced with various modified ribonucleotides. Curlcake refers to publish data from Liu et. al. (9). A significantly lower quality of modified IVT RNA (cyan) was obtained compared with nonmodified IVT RNA (blue) and cDNA (red) with *** P < e−100. (D) Characteristic of RNA rBEMs obtained with pentamers (k5) and heptamers (K5+2). The density plot shows marginal rug on the bottom of maximum %ESB values of individual kmers. Boxplots include %ESB across positions producing the highest maximum %ESB of rBEM_k5 (green) and rBEM_k5+2 (magenta). These were all related to homopolymers of C or G. (IVT, in vitro transcription; %ESB, percentage Error of Specific Base; dRNAO, modified mRNA; dcDNAO, corresponding cDNA by direct sequencing; dRNAU, RNA produced with unmodified uridine by direct sequencing; rBEMs, RNA background error models).
To illustrate the effect on recorded signals when modified bases are present, in Figure 1B the re-squiggled signals are compared for a small region (position 989–1009) of the luciferase gene containing four U bases in three positions. The sequence signals obtained with dcDNAO (Figure 1B, red in top panel) or from dRNAU (Figure 1B, blue in bottom panel) matched those of the theoretical canonical signal model for DNA. In contrast, the re-squiggled signals of dRNAO containing modified U (Figure 1B, cyan in bottom panel) were altered compared to the canonical RNA signals. Thus, the presence of 5moU bases in the RNA template most likely caused some of the observed perturbations, while a RT step to produce cDNA removed this effect. The 5moU sites and the bases in their vicinity produced dramatically perturbed signals in dRNAO, as is clearly visible for a C at position 997 (Figure 1B, bottom panel). This has a direct impact on the accuracy of base calling. Note that base calling is typically performed on a window of pentamers (19) so that any effect due to the presence of a modified base can affect the signal of bases in its direct vicinity.
The positions for which %ESB exceeded the cutpoint of 25% were recorded for the complete dRNAO template and for the dRNAU and dcDNAO templates (Supplementary Figure S1). High %ESB values were more frequently obtained with the dRNAO template than with either the dRNAU or the dcDNAO. Further, in positions where 5moU was present, a higher %ESB was frequently produced. We also recorded greater than 25% ESB values for some positions where other bases were present, while not all positions with 5moU increased the %ESB in the dRNAO reads. Some of the observed errors derived from translocation through the nanopore and ionic current alterations caused a glitch in the corresponding output. In a number of cases, a high %ESB coincided with the presence of homopolymeric stretches. Although these signals are not easily distinguishable from base modifications signals, homopolymeric stretches can be readily identified from the sequence. Further, elevated %ESB values observed in both dRNAU and dcDNAO are more likely to be caused by structural features irrespective of the presence of modified bases as systematic background noises derived from the base-calling algorithm that can be modeled.
Next, we compared the read mean quality score, which reflected the sequencing error derived from incorrect interpretation of base calling of IVT RNAs with modified bases versus corresponding unmodified IVT RNA derived from synthetically constructed DNA templates (see Methods). A similar sequence dataset containing m6A called Curlcake, from a previous study (9), was also included. Apart from 5moU and m6A, we investigated other modified ribonucleosides known to be present in mRNA (33–35) including m1A, m5C, hm5C, f5C, m7G, Ino and psU. The presence of each of these modified bases significantly reduced the read mean quality score (P < 1e−100) (Figure 1C).
To systematically capture the background noise related to the presence of a given pentamer, we constructed rBEMs of all pentamers (rBEM_k5 and an extension of two nucleotides at the 3′-end (rBEM_k5+2) to capture the impact of longer homopolymers, from nonmodified IVT RNA sequences derived from the human transcriptome. The distribution of the obtained %ESB values of rBEMs is shown in the graph of Figure 1D and the distribution of all individual kmer occurrences is provided in Supplementary Figure S2. In total, rBEM_k5 consists of 1,024 pentamers and rBEM_k5+2 consists of 16,083 heptamers, of which 301 heptamers were not found in the human transcriptome. The histogram illustrates a variation of the background error among individual kmers that appear to reflect sequence-context–dependent behavior. Although a similar distribution of background errors was obtained with both rBEMs, the rBEM_k5+2 captured slightly more background errors due to the longer sequence length. As expected, homopolymer stretches of G5 and C5 produced the highest background error for rBEM_k5. The impact of background errors due to the homopolymers is even stronger in rBEM_k5+2. The constructed rBEMs were used as references for the identification of RNA modifications on the native RNA sequences using the developed ELIGOS software.
Sequencing errors of native RNA can predict common RNA modifications but not m5C
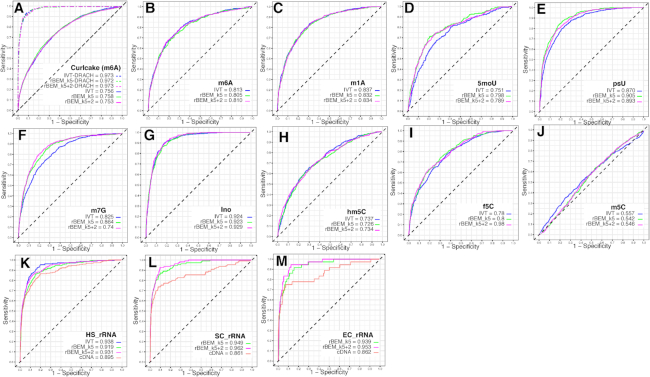
We next evaluated the RNA modification prediction performance of ELIGOS by using rBEMs as the reference and the modified IVT RNA datasets that contained nine types of base modifications (m6A, m1A, 5moU, psU, m7G, Ino, hm5C, f5C and m5C). The results were compared to the optimal reference of nonmodified IVT RNA, which represents the best reference but may not always be available, and to the cDNA (Figure 2A–L). We used the odds ratios, which represent the level of error of native sequence over the unmodified RNA or rBEMs, as the predictor of sites with potential modifications. Because the presence of a methylated base can influence the differential %ESB of adjacent positions as seen in Figure 1B, flanking bases should also be considered. Thus, we evaluated the performance of RNA modification predictions by ELIGOS by statistical tests for three scenarios: (i) the effect was only calculated at the position where the modification was present; (ii) the effect was extended to one position on both sides of the modified base position; (iii) the effect was extended with two positions on both sides of the modified base position. Based on odd ratio values presented in Supplementary Figure S3, we found that these different scenarios produced different prediction performances, depending on the type of RNA modifications that is present. For example, the third scenario gave the best prediction performance on the m7G dataset, but the first scenario gave the best performance on the Ino dataset. This indicates that, as expected, the sequence context and the type of modification affects the signal alteration of modified and adjacent positions. In general (with the exception m5C), using maximum odd ratios among the three scenarios for individual positions gave the best consistency performance for predicting RNA modifications (AUC = 0.73–0.93). Both rBEMs resulted in a similar prediction performance in the synthetic datasets, with comparable results, when using nonmodified IVT RNA as the reference sequence. Interestingly, the prediction performance of m5C was poor, even though we observed a similar trend of low read mean quality scores as with other RNA modifications. This indicated that signal alteration by m5C is not strong enough to alter the Guppy base caller outcome. This might be due to m5C being such a common modification that Guppy software does not distinguish between C and m5C and call both as C. Unfortunately, we found that ELIGOS cannot reliably predict the presence of m5C at this time. Next, we investigated ELIGOS prediction performance for a known biological context, the DRACH (D = G/A/U, R = G/A, H = A/U/ C) motif-containing m6A in the Curlcake dataset (9). This resulted in excellent prediction accuracy (AUC > 0.97; Figure 2A and Supplementary Figure S3B, D) that was comparable with the results reported by Liu et al. (9).
Figure 2.
ROC curve plot with AUC values representing the prediction performance of RNA modification using ELIGOS in synthetic modified IVT RNA datasets as illustrated in Figure 1C and rRNA dataset of the three model organisms. (A) published m6A Curlcake dataset. The dashed lines indicate ROC curves when considering the DARCH motif (9). (B) m6A. (C) m1A. (D) 5moU. (E) psU. (F) m7G. (G) hm5C. (H) m5C. (I) f5C. (J) m5C. (K) rRNA of human cells (HS). (L) rRNA of yeast (SC) and (M) rRNA of E. coli (EC). Blue, red, green and magenta lines represent the prediction performance when use nonmodified IVT RNA, cDNA, rBEM_k5 and rBEM_k5+2 as the reference, respectively. The selected best performance for each dataset was used for the plots. The details of all prediction scenarios are provided in Supplementary Figure S3. (ROC, Receiver Operating Characteristic; AUC, Area Under Curve; IVT, in vitro transcription; (as B to M are explained above); rBEMs, RNA background error models)
Accurate prediction of RNA modification on rRNA molecules using ELIGOS
We continued our investigations with naturally modified RNAs, for which the rRNA from E. coli, yeast, and human cell lines were analyzed. These RNA molecules are naturally modified but contain much lower fractions of modified ribonucleotides than the IVT RNA molecules. We observed an increase of %ESB of the native rRNAs sequences due to modification when compared with corresponding cDNA sequences (Supplementary Figure S4). The results of the ROC curve analysis on the rRNAs are shown in Figure 2K–M. The performance of the predictions was benchmarked against experimentally validated RNA modifications on the bases and sugar moieties of the rRNAs, as was recently described by Taoka et al. (36). For the rRNA of human cell lines (Figure 2I), it is obvious that using rBEMs derived from IVT RNA produced from the human transcriptome as the reference produced comparable results to the reference of nonmodified IVT RNA (AUC > 0.91), while satisfactory results were obtained for the other two organisms, indicating the robustness of the method. The prediction performance using cDNA produced slightly lower AUC values than rBEMs for the three rRNA datasets, possibly due to the differences in background derived from the sequencing chemistry of a DNA molecule that has an opposite orientation (5′ to 3′ for dcDNA-seq and 3′ to 5′ for dRNA-seq). Moreover, DNA passes the pore at a higher speed (450 nt/s for dcDNA-seq) than RNA (70 nt/s for dRNA-seq) during sequencing. The prediction performance for the rRNAs of rBEM_k5+2 was slightly better than rBEM_k5 because the first better captures the background error derived from long homopolymer sequences (Supplementary Figure S5).
In summary, we were able to capture most of the common RNA modifications found in nature in various RNA sequencing datasets, including sequences generated with synthetic IVT RNA and rRNA of three species using ELIGOS. This software can accurately detect a variety of modified ribonucleotides simultaneously in sequences obtained from native RNA, with the exception of m5C. Notably, as rBEM_k5+2 produced results that were comparable with nonmodified IVT RNA and better than rBEM_k5, we conclude that using rBEM_k5+2 data as a reference can give satisfactory results without the need to perform addition IVT experiments.
Uncovering known RRACH motif from m6A in yeast during meiosis state in vivo
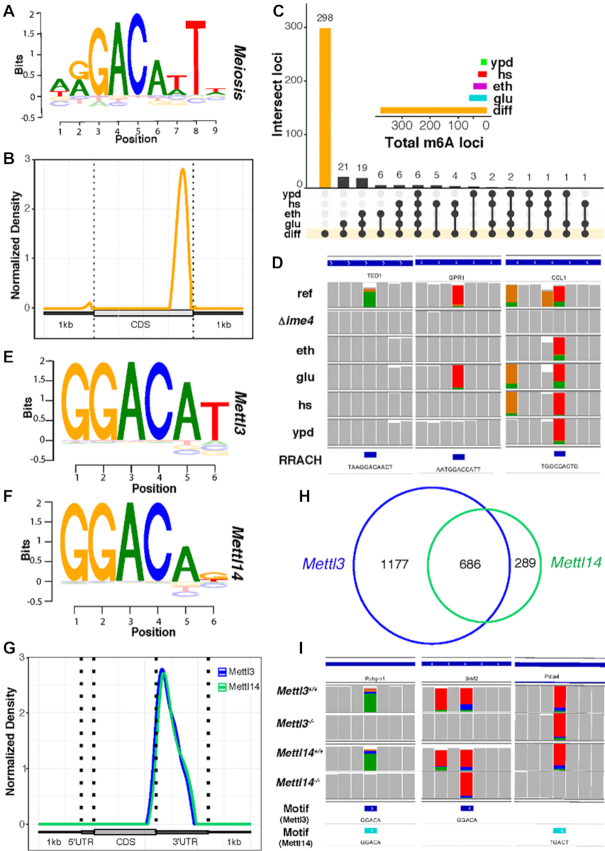
Next, we demonstrated the capability of ELIGOS by performing epitranscriptional landscapes analysis in vivo of the well-study RNA modification m6A. We applied ELIGOS on the published dataset (9) of the yeast reference and Δime4 knockout, producing m6A free transcripts under meiosis state (37). By this experimental setup, we can identify differential m6A by direct comparison of the native transcript sequences between the reference strain to the knockout strain. As previously reported, the median level of m6A modifications of the reference strain is around 20% (38); therefore, an odds ratio of 1.2 with P < 0.001 used as the cut-off for identification of differential m6A resulted in 1,513 locations, consisting of 736 sites for A, 249 sites for C, 235 sites for G, and 293 sites for U. We then extracted 6 bases surrounding the identified A sites, considering the differential m6A sites and used as the input to BaMM software (21) to identify the consensus motif. With the unbiased procedure, we uncovered an m6A consensus sequence, which is almost identical to the RRACH motif, previously reported for yeast, using the m6A-seq method (WRG-m6A-CAWTW) (37) (Figure 3A). We scanned the consensus motif (Figure 3A) back to the extracted sequences by BaMM software to identify high confidence m6A methylated positions that resulted in 392 positions that strongly bias the 3′ end of the transcripts (Figure 3B), which is in agreement with a previous report (37).
Figure 3.
Differential epitranscriptional analysis of native transcriptome sequences derived from yeast (A–D) and mESCs (E-I) using ELIGOS. (A) Sequence logo plot of uncovered RRACH motif from differential adenine sites derived from the yeast meiosis dataset. (B) Standardized transcript coordinate plot of normalized density derived from the transcripts containing the RRACH motif to illustrate its preferential position in 3′ untranslated regions. (C) Upset plot shows the presence/absence of the 392 high confidence positions of the RRACH motif across different growth conditions (diff, the high confidence position; ypd, rich media; hs, rich media with heat shock treatment; glu, glucose limited minimal media; eth, ethanol limited minimal media). The insert shows the distribution of the 392 high confidence position across different growth conditions. (D) Integrative Genomics Viewer(IGV) snapshot with the gene name below the sequence of the three examples of differential m6A position of meiosis specific (left), common between meiosis and starvation on glucose limited growth (middle), and common among meiosis and other growth condition (right). The plot shows across different growth conditions (ref, miosis state of reference strain; Δime4, miosis state of ime4 knockout strain). The bottom row shows the location of RRACH like motif. (E) Logo plot of DRACH like motif from differential adenine sites derived from mESCs Mettl3 dataset. (F) Logo plot of DRACH like motif from differential adenine sites derived from mESCs Mettl14 dataset. (G) Standardized transcript coordinate plot of normalized density derived from the identified motif of Mettl3 dataset (blue) and Mettl14 dataset (floral green). (H) Venn diagram shows the comparison of the identified DRACH like motif positions between Mettl3 dataset (blue) and Mettl14 dataset (floral green). (I) IGV snapshot with the gene name below the sequence of the three examples of differential m6A positions of common positions (left) and specific positions for Metl13(middle) and Metl14(right) dataset. The last two rows show the location of DRACH-like motif derived from the two mESCs datasets.
To evaluate whether the identified epitranscriptional regulation of m6A position is meiosis state-specific, we performed a comparison of the high confidence m6A position with the transcriptome data from different growth conditions such as carbon limited minimal media of glucose (glu) (6), ethanol (eth) (6), rich media (ypd), and rich media with heat shock (hs) treatment. The results derived from ELIGOS analysis of those transcriptomes using rBEM_k5+2 as the reference was calculated at the considered position (scenario 1); this gave a good prediction performance on m6A modifications (Supplementary Figure S3 panel E) to avoid the impact of other RNA modifications nearby. The same cut-off of odds ratio of 1.2 with P < 0.001 was used for the comparison. The upset plot (Figure 3C) shows the dynamic of m6A during various growth phases and growth conditions, indicating meiosis-dependent regulation of m6A as reported previously (39). We detected approximately 20 positions of m6A for carbon starvation growth conditions (glu, eth), likely due to the common m6A regulation in the meiosis and starvation pathway (39). The selected examples of growth condition-dependent m6A regulated positions (Figure 3D) on transcripts of TED1 for meiosis-dependent, GPR1 for common between meiosis and starvation on glucose-limited growth, and CCL1 for common among meiosis and the other growth conditions, increased sequencing errors due to the presence of m6A.
Uncovering known DRACH-like motif from m6A in mESCs in vivo
We further demonstrated the capability of ELIGOS in mESCs, which have m6A methyltransferase complex of METTL3 and METTL14 as the key cellular machinery for m6A regulation. We performed native RNA sequencing on the Mettl13 and Mettl14 knockout mESCs and their reference and used the data to identify differential RNA methylation sites among them by comparing the reference cells with the knockout cells similarly to the yeast dataset. When comparing Metl13 knockout with the reference, and using the same statistical cut-off as the yeast dataset, we identified 10,569 differential sites consisting of 43% of sites for A, 18% for C, 18% for G, and 22% sites for U. For the Mettl14 knockout dataset, we identified 3,110 differential sites consisting of 52% of sites for A, 15% for C, 12% for G and 21% sites for U base. We then extracted 6 bases surrounding the identified A sites from both datasets and used it as the input to BaMM discovery software to identify the consensus motif. With the unbiased procedure, we uncovered consensus motifs that were similar to the known canonical m6A methylation DRACH motif (40,41), and found in both of Mettl13 (Figure 3E) and Mettl14 (Figure 3F) knockout dataset. We next extracted and analyzed the position of the identified consensus motifs along with each transcript presented in a standardized coordinate plot (Figure 3G). This identified a clear preference for the DRACH-like motif to be present at the gene-bordering flank of the 3′ untranslated region (UTR), which agrees with previous studies (40,42–44). We compared the identified consensus motif-containing position between the two datasets (1,863 for the Mettl13 dataset and 975 for the Mettl14 dataset) and found a high overlap among them as illustrated in the Venn's diagram in Figure 3H. This indicates the synergic activity of the m6A methyltransferase function of Mettl13 and Mettl14 on mRNA as reported previously (45,46). The selected examples of positions in the transcripts of Pabpn1, Srsf2 and Pdia4 represent a common m6A position and specific positions for Mettl13 and Mettl14 datasets (Figure 3I).
Epitranscriptional analysis on human transcriptome using ELIGOS
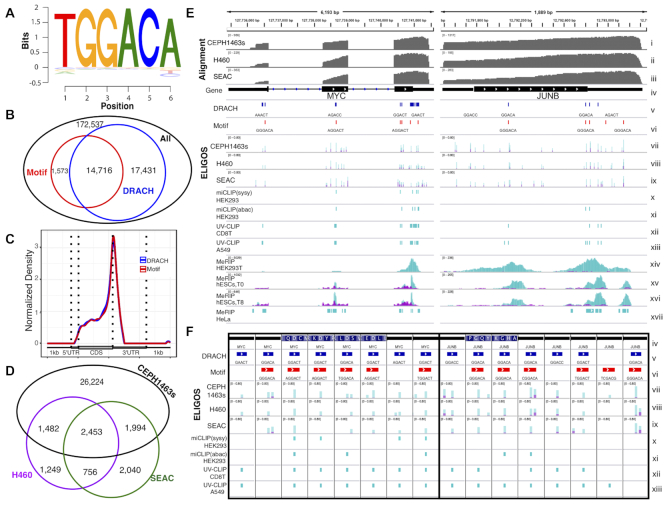
Lastly, we analyzed the transcriptome of the reference native RNA sequencing dataset of a transcriptome derived from the human cell line CEPH1463 (7) using the rBEM_k5+2 as the reference. We used a stringent cut-off of the odds ratio of 2.5 and adjusted P < 10e–5 to identify RNA modification sites. Based on the cut-off, 1,039,699 sites were identified with 210,966 sites for A. We then extracted 6 bases surrounding the identified A sites from the dataset and used them for the BaMM software, determined the consensus motif, and established corresponding locations on the transcripts (Figure 4A). The consensus motif has pattern similar to the known DRACH for m6A modification (40,41) with an additional T base on the 5′ end and a high probability of G based on positions 2 and 3, indicating higher pattern specificity. We compared the locations of the consensus motif with the locations of the identified A sites on the DRACH motif sequence (Figure 4B) and found a high fraction overlap among them. The 1,573 nonoverlapping positions with a DRACH sequence pattern were observed due to the relaxing criteria of the motif scan algorithm of BaMM software. The identified motif (Figure 4A) was the most abundant subset motif of the DRACH sequence pattern (Supplementary Table S1). We next analyzed the positions of the identified A sites with the DRACH motif and the consensus DRACH-like motif along with each transcript and graphed it in a standardized coordinate plot of normalized density (Figure 4C). This identified a clear preference for the motifs to be present at the gene-bordering flank of the 3′ UTR, which agrees with previous studies (40,42–44). Therefore, the identified A sites with the DRACH motif could be considered as m6A. We evaluated the impact of the cellular mutations on the m6A profiling results by identifying point mutations from cDNA sequences using Li's method (47). We found that less than 0.35% of the identified DRACH motif contained point mutations (Supplementary Figure S6). However, the detected point mutations from cDNA sequences can be derived from missed-base incorporation of RT due to the presence of some modifications on the mRNA template (48), e.g. Ino can be recognized as G by RT, then C is incorporated into the cDNA as recently reported in ONT by Workman et al. (7). Therefore, genome resequencing may be required for construction of an accurate reference genome for the RNA modification profiling.
Figure 4.
Epitranscriptional landscape analysis of human cells using ELIGOS. (A) Sequence logo plot of the identified consensus DRACH like motif surrounding differential A identified by ELIGOS from the CEPH1463 dataset (7). (B) Venn diagram showing a comparison between the identified position of DRACH-like motif from panel A (red) with position of the DRACH (blue) of the differential %ESB adenine sites (black). (C) Standardized transcript coordinate plot of normalized density of transcripts containing the identified consensus motif (red) and DRACH motif with identified differential A sites (blue) to illustrate its preferential position in 3′ untranslated regions. (D) Venn's diagram shows the comparison of differential %ESB adenine sites surrounding the DRACH motif sequence for the three human cells CEPH1463 (black), H460 (magenta) and SAEC (green). (E) Examples of selected oncogene transcripts of MYC (left panel) and JUNB (right panel) in which both the identified consensus DRACH-like motif and the DRACH motif were found to be modified. A comparison is shown in IGV Genome Browser of our predictions and previous studies conducted with different human cells and different m6A profiling methods. The tracks show (from top down): alignment coverage depth of dRNA reads of the transcripts derived from CEPH1463 (i), H460(ii) and SAEC (iii) dataset; (iv) transcript architecture; (v) location of the DRACH motifs (blue); (vi) location of the identified consensus DRACH-like motifs (red); overlay bar plot of %ESB of native RNA sequences (cyan) and rBEM_k5+2 (magenta) at the differential %ESB positions for A as identified by ELIGOS for the three human cells CEPH1463 (vii), H460 (viii) and SAEC (ix); (x) m6A miCLIP data of HEK293 cells using SySy m6A antibody enrichment; (xi) miCLIP data of HEK293 cells using Abacam m6A antibody enrichment; (xii) UV crosslinking and immunoprecipitation (UV-CLIP) data of CD8T cells; (xiii) UV-CLIP data of A549 cells; (xiv) MeRIP peak data of HEK293T cells; (xv) MeRIP peak data of hESCs cells at time point T0; (xvi) MeRIP peak data of hESCs cells at time point T48. All MeRIP peak data were plotted based on the read coverage depth of m6A enriched (cyan) and the reference sequencing library (magenta); (xvii) MeRIP peak region data of HeLa cells. (F) A zoomed output comparison shows single base resolution for m6A positions identified by ELIGOS agreed with miCLIP and UV-CLIP methods. The track information is the same as in panel E).
RNA methylation, especially m6A, plays an important role in carcinogenesis and treatment response (49,50), including lung cancer (7). We performed additional pilot experiments of native RNA sequencing on lung cancer cell line H460 and primary SAEC to compare m6A profile with the CEPH1463s. With the same cut-off, 379,882 sites were identified for H460 with 75,428 sites for A, and 319,353 sites were identified for SAEC cells with 59,291 sites for A. Approximately 10% of the identified A sites located on the DRACH motif sequences (5,940 for H460 and 7,243 SAEC cells) were considered to be m6A sites. Note that the number of identified modification sites of the lung cells was much less than the CEPH1463 cells due to the much lower sequencing depth in the H460 and SAEC cells compared to the CEPH1463 cells. We compared the identified m6A sites among the three cells presented in a Venn diagram, in Figure 4D. The H460 and SAEC had 3,209 overlapped m6A sites with 2,731 unique m6A sites for H460 and 4,034 unique m6A sites for SAEC cells. Most of the m6A modification sites identified in lung cells were in common with the CEPH1463 cells. Even though lung cell datasets have lower sequencing depth, we observed over 4,045 m6A sites that were identified in the lung cells and not identified in the CEPH1463 cell. These could indicate the cell type-specific regulation of m6A RNA modification.
We investigated the ELIGOS results of m6A with other published methods with two oncogenes transcript, MYC and JUNB, and presented in IGV snapshot (Figure 4E). For MYC transcript, ELIGOS identified that the m6A position (identified A sites with DRACH motif sequences) was mostly consistent with the UV cross-linking immunoprecipitation (UV-CLIP) method (42) and methylated RNA immunoprecipitation (MeRIP) data of HeLa cells (51). We observed the absence of an m6A site on the 5′-UTR in the m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP) data of HEK293 cells (40), in MeRIP data from HK239T cells (43), and in MeRIP data from hESCs (40). For the JUNB transcript, the results from the MeRIP data of HK239T (43) and hESCs (31) seem to have the most consistency with ELIGOS results. On the other hand, miCLIP(abacam) data of HEK293 cells fail to identify all of m6A on the JUNB transcript.
The inconsistency of m6A detection across different studies indicates highly complex and dynamic cellular regulation of methylation patterns that is cell type-specific and method dependent. The miCLIP and UV-CLIP can give single-base resolution for m6A identifications, so we focused on the DRACH containing position of the two transcripts (Figure 4F) to evaluate the ELIGOS result. ELIGOS correctly identified the m6A position at single-base resolution, and agreed with the miCLIP and UV-CLIP methods.
DISCUSSION
The major fraction of sequencing errors by ONT, which captures single-molecule sequences, is derived from stochastic noise that can be corrected by consensus base calling from reads pile-up (52). The consensus error correction approach typically results in the correction of sequencing errors when DNA is sequenced; however, approximately 1% of the total errors typically needs to be further polished by short reads (52). The sequencing of native RNA results in more errors, as we found higher %ESB scores for this template (Figure 1A).
When present, base modifications of nucleic acids alter the ionic current signal recorded during ONT sequencing, leading to errors that are inherent to the application of the helicase and the pore protein for passage through the pore. We developed ELIGOS for determining a comparative error analysis of long-read sequences, as this can be used as a signature to recognize base modifications. By sequencing IVT RNA, we can compare the errors recorded with modified RNA to that of nonmodified RNA or cDNA signals. The use of native RNA sequences from nonmodified RNA obtained by IVT as a reference is suitable to eliminate systematic errors. Nevertheless, the construction of IVT to study genome-wide RNA modifications is not trivial. Therefore, we developed the rBEMs that can mimic error profiles across different sequence contexts representing the systematic error of dRNA-seq. Using rBEM as the reference to identify RNA modification from native RNA sequences gave a comparable prediction performance with nonmodified RNA in synthetic modified RNA datasets and rRNA datasets. Moreover, ELIGOS captured most of the known RNA methylation sites, for all four bases simultaneously, despite inherent differences in methylation of these bases or the sugar backbone. This was demonstrated in E. coli, yeast, and human RNA. This provides a promising approach to detect expected and novel RNA methylations and base modifications directly from native RNA sequences. This capability is superior to traditional methods that can detect only one type of methylation at a time and require complex experimental procedures. Moreover, based on the same principle, ELIGOS can be applied to identify DNA modifications by the comparison of the errors between native DNA and cDNA or a PCR product (Supplementary Figure S7). This potential will need to be further investigated and compared with existing methods for direct DNA modification detection using ONT (20,53) or PacBio (54) sequencing (Pacific Biosciences, Menlo Park, CA).
Understanding the RNA modification regulation of RNA methyltransferase can be accomplished by comparative investigation between the wild type cell and specific inactivation of the RNA methyltransferase through gene knockout or knockdown. Differential %ESB analysis between wild type and m6A methylase knockout using ELIGOS was applied in yeast and mammalian cell system to assess the impact of the m6A methyltransferase. With ELIGOS, we can unbiasedly uncover the known biological importance of the RRACH/DRACH motifs directly from the analysis of differential %ESB sites. Using the same strategy of differential %ESB analysis, using ELIGOS to study different RNA methyltransferase will improve our understanding of cellular regulation of other RNA methyltransferase and their role in the development and disease processes.
Using ELIGOS to identify epitranscriptional landscapes by mean of rBEM_k5+2, we were able to uncover known biologically relevant motifs containing m6A RNA in human cell datasets. We found that approximately 10% of the identified A sites could be m6A sites, based on their sequence context of the DRACH motif. The rest of the A sites could be other types of modifications of A such as m1A as previously reported (55,56). The identified modification sites of the other bases will need further investigations to uncover such known (57) and novel ribonucleotide modifications in mRNA.
ELIGOS can specifically identify the location of RNA modifications, but at this time it cannot tell the exact type of RNA methylation because sequencing errors are used as the proxy of detection. This limitation requires further investigations to determine the nature of the RNA modification position inferred by ELIGOS, by using traditional techniques such as the LC–MS/MS approach (16). Alternatively, signal level analysis coupled with machine learning in the future might be able to discern the modification types from the ONT signal, as recently demonstrated on DNA modification (58). Besides, the input data for our method depend on the results obtained from base calling and long-read aligner software as a prerequisite. Therefore, the accuracy of these steps will influence the final result, and as they improve, so will the ELIGOS results. Lastly, it is possible that the method may be over-reporting the number of predicted modified bases due to the noisy nature of the ONT outputs.
In conclusion, this study provides a concrete foundation to study native RNA sequences that carry important information on RNA modifications. Detailed investigations to dissect the complex properties of RNA from detected error signatures is now feasible. Our ELIGOS software is publicly available and can be used to detect possible RNA modification sites quickly and on a global transcriptomic scale. The study generated rich native RNA sequencing datasets of various synthetic modified RNA, rRNAs, and the transcriptome of mouse and human cell lines that is useful for the research community in advancing the development of bioinformatics software to analyze such data. Moreover, ELIGOS can be used as a diagnostics tool to improve the base-calling algorithm of nanopore sequencing. We envisage that the sequencing of native RNA will become a powerful and versatile tool to advance RNA biology.
DATA AVAILABILITY
All data generated fastq data in this study were deposited in the SRA database (accession number SRP166020). ELIGOS is available at https://gitlab.com/piroonj/eligos2.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: I.N. designed and conceived the project. P.J. developed and implemented ELIGOS software. T.W. and T.D.W. performed MinION sequencing for dRNA-Seq and dcDNA-Seq and prepared the data for submission. T.W. and T.D.W. perform in vitro transcription, yeast, and E. coli experiments. N.S.A. performed human cell line experiments. J.L. perform mESCs experiments. P.J. and I.N. performed computational analysis and with T.M.W. interpreted the data. D.U., A.T.F., G.B., Q.D., M.L.J. and C.H. participated in the study design. I.N., T.W., T.M.W., P.J. and T.D.W. wrote and revised the manuscript. All authors have read and approved the final version.
Notes
Present address: Aime T. Franco, Division of Endocrinology and Diabetes, The Children’s Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Contributor Information
Piroon Jenjaroenpun, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Thidathip Wongsurawat, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Taylor D Wadley, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Trudy M Wassenaar, Molecular Microbiology and Genomics Consultants, Zotzenheim, Germany.
Jun Liu, Department of Chemistry, Department of Biochemistry and Molecular Biology, Howard Hughes Medical Institute, The University of Chicago, Chicago, IL 60637, USA.
Qing Dai, Department of Chemistry, Department of Biochemistry and Molecular Biology, Howard Hughes Medical Institute, The University of Chicago, Chicago, IL 60637, USA.
Visanu Wanchai, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Nisreen S Akel, Department of Physiology and Biophysics, College of Medicine, The University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Azemat Jamshidi-Parsian, Department of Radiation Oncology, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Aime T Franco, Department of Physiology and Biophysics, College of Medicine, The University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Gunnar Boysen, Department of Environmental and Occupational Health, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Michael L Jennings, Department of Physiology and Biophysics, College of Medicine, The University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
David W Ussery, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Chuan He, Department of Chemistry, Department of Biochemistry and Molecular Biology, Howard Hughes Medical Institute, The University of Chicago, Chicago, IL 60637, USA.
Intawat Nookaew, Department of Biomedical Informatics, College of Medicine, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA; Department of Physiology and Biophysics, College of Medicine, The University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Helen Adams and Arkansas Research Alliance Endowed Chair; Arkansas Biosciences Institute; National Institute of General Medical Sciences of the National Institutes of Health [P20GM125503 to I.N.] (in part); National Human Genome Research Institute [RM1 HG008935 to C.H.]; CH is a Howard Hughes Medical Institute Investigator. Funding for open access charge: UAMS internal funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mutz K.O., Heilkenbrinker A., Lonne M., Walter J.G., Stahl F.. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2013; 24:22–30. [DOI] [PubMed] [Google Scholar]
- 2. Nookaew I., Papini M., Pornputtapong N., Scalcinati G., Fagerberg L., Uhlen M., Nielsen J.. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 2012; 40:10084–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carrara M., Beccuti M., Lazzarato F., Cavallo F., Cordero F., Donatelli S., Calogero R.A.. State-of-the-art fusion-finder algorithms sensitivity and specificity. Biomed. Res. Int. 2013; 2013:340620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hansen K.D., Brenner S.E., Dudoit S.. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010; 38:e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garalde D.R., Snell E.A., Jachimowicz D., Sipos B., Lloyd J.H., Bruce M., Pantic N., Admassu T., James P., Warland A. et al.. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods. 2018; 15:201–206. [DOI] [PubMed] [Google Scholar]
- 6. Jenjaroenpun P., Wongsurawat T., Pereira R., Patumcharoenpol P., Ussery D.W., Nielsen J., Nookaew I.. Complete genomic and transcriptional landscape analysis using third-generation sequencing: a case study of Saccharomyces cerevisiae CEN.PK113-7D. Nucleic Acids Res. 2018; 46:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Workman R.E., Tang A.D., Tang P.S., Jain M., Tyson J.R., Razaghi R., Zuzarte P.C., Gilpatrick T., Payne A., Quick J. et al.. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods. 2019; 16:1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Smith A.M., Jain M., Mulroney L., Garalde D.R., Akeson M.. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One. 2019; 14:e0216709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu H., Begik O., Lucas M.C., Ramirez J.M., Mason C.E., Wiener D., Schwartz S., Mattick J.S., Smith M.A., Novoa E.M.. Accurate detection of m(6)A RNA modifications in native RNA sequences. Nat. Commun. 2019; 10:4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crecy-Lagard V., Ross R., Limbach P.A., Kotter A et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F.. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011; 39:D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xuan J.J., Sun W.J., Lin P.H., Zhou K.R., Liu S., Zheng L.L., Qu L.H., Yang J.H.. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018; 46:D327–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saletore Y., Meyer K., Korlach J., Vilfan I.D., Jaffrey S., Mason C.E.. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012; 13:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He C. Grand challenge commentary: RNA epigenetics. Nat. Chem. Biol. 2010; 6:863–865. [DOI] [PubMed] [Google Scholar]
- 15. Helm M., Motorin Y.. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 2017; 18:275–291. [DOI] [PubMed] [Google Scholar]
- 16. Jonkhout N., Tran J., Smith M.A., Schonrock N., Mattick J.S., Novoa E.M.. The RNA modification landscape in human disease. RNA. 2017; 23:1754–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rang F.J., Kloosterman W.P., de Ridder J.. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018; 19:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wick R.R., Judd L.M., Holt K.E.. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019; 20:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W.. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017; 14:407–410. [DOI] [PubMed] [Google Scholar]
- 21. Kiesel A., Roth C., Ge W., Wess M., Meier M., Soding J.. The BaMM web server for de-novo motif discovery and regulatory sequence analysis. Nucleic Acids Res. 2018; 46:W215–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R.. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010; 11:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conway J.R., Lex A., Gehlenborg N.. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017; 33:2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cui X., Wei Z., Zhang L., Liu H., Sun L., Zhang S.W., Huang Y., Meng J.. Guitar: an R/Bioconductor package for gene annotation guided transcriptomic analysis of RNA-related genomic features. Biomed. Res. Int. 2016; 2016:8367534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sachs M.C. plotROC: a tool for plotting ROC curves. J Stat Softw. 2017; 79:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wongsurawat T., Jenjaroenpun P., Taylor M.K., Lee J., Tolardo A.L., Parvathareddy J., Kandel S., Wadley T.D., Kaewnapan B., Athipanyasilp N. et al.. Rapid sequencing of multiple RNA viruses in their native form. Front Microbiol. 2019; 10:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Che S., Ginsberg S.D.. Amplification of RNA transcripts using terminal continuation. Lab. Invest. 2004; 84:131–137. [DOI] [PubMed] [Google Scholar]
- 30. Tang G.Q., Bandwar R.P., Patel S.S.. Extended upstream A-T sequence increases T7 promoter strength. J. Biol. Chem. 2005; 280:40707–40713. [DOI] [PubMed] [Google Scholar]
- 31. Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K. et al.. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014; 15:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sappington D.R., Siegel E.R., Hiatt G., Desai A., Penney R.B., Jamshidi-Parsian A., Griffin R.J., Boysen G.. Glutamine drives glutathione synthesis and contributes to radiation sensitivity of A549 and H460 lung cancer cell lines. Biochim. Biophys. Acta. 2016; 1860:836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roundtree I.A., Evans M.E., Pan T., He C.. Dynamic RNA modifications in gene expression regulation. Cell. 2017; 169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paul M.S., Bass B.L.. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998; 17:1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang H.Y., Xiong J., Qi B.L., Feng Y.Q., Yuan B.F.. The existence of 5-hydroxymethylcytosine and 5-formylcytosine in both DNA and RNA in mammals. Chem. Commun. (Camb.). 2016; 52:737–740. [DOI] [PubMed] [Google Scholar]
- 36. Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N et al.. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018; 46:9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwartz S., Agarwala S.D., Mumbach M.R., Jovanovic M., Mertins P., Shishkin A., Tabach Y., Mikkelsen T.S., Satija R., Ruvkun G. et al.. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell. 2013; 155:1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garcia-Campos M.A., Edelheit S., Toth U., Safra M., Shachar R., Viukov S., Winkler R., Nir R., Lasman L., Brandis A. et al.. Deciphering the “m(6)A Code" via antibody-independent quantitative profiling. Cell. 2019; 178:731–747. [DOI] [PubMed] [Google Scholar]
- 39. Agarwala S.D., Blitzblau H.G., Hochwagen A., Fink G.R.. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLos Genet. 2012; 8:e1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patil D.P., Pickering B.F., Jaffrey S.R.. Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 2018; 28:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ke S., Alemu E.A., Mertens C., Gantman E.C., Fak J.J., Mele A., Haripal B., Zucker-Scharff I., Moore M.J., Park C.Y. et al.. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015; 29:2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang C., Chen Y., Sun B., Wang L., Yang Y., Ma D., Lv J., Heng J., Ding Y., Xue Y. et al.. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017; 549:273–276. [DOI] [PubMed] [Google Scholar]
- 45. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al.. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C.. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014; 16:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011; 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Potapov V., Fu X., Dai N., Correa I.R. Jr, Tanner N.A., Ong J.L.. Base modifications affecting RNA polymerase and reverse transcriptase fidelity. Nucleic Acids Res. 2018; 46:5753–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thapar R., Bacolla A., Oyeniran C., Brickner J.R., Chinnam N.B., Mosammaparast N., Tainer J.A.. RNA modifications: reversal mechanisms and cancer. Biochemistry. 2019; 58:312–329. [DOI] [PubMed] [Google Scholar]
- 50. Delaunay S., Frye M.. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019; 21:552–559. [DOI] [PubMed] [Google Scholar]
- 51. Chen K., Lu Z., Wang X., Fu Y., Luo G.Z., Liu N., Han D., Dominissini D., Dai Q., Pan T. et al.. High-resolution N(6) -methyladenosine (m(6) A) map using photo-crosslinking-assisted m(6) A sequencing. Angew. Chem. Int. Ed. Engl. 2015; 54:1587–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Senol Cali D., Kim J.S., Ghose S., Alkan C., Mutlu O.. Nanopore sequencing technology and tools for genome assembly: computational analysis of the current state, bottlenecks and future directions. Brief. Bioinform. 2018; 20:1542–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rand A.C., Jain M., Eizenga J.M., Musselman-Brown A., Olsen H.E., Akeson M., Paten B.. Mapping DNA methylation with high-throughput nanopore sequencing. Nat. Methods. 2017; 14:411–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Flusberg B.A., Webster D.R., Lee J.H., Travers K.J., Olivares E.C., Clark T.A., Korlach J., Turner S.W.. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods. 2010; 7:461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou H., Rauch S., Dai Q., Cui X., Zhang Z., Nachtergaele S., Sepich C., He C., Dickinson B.C.. Evolution of a reverse transcriptase to map N(1)-methyladenosine in human messenger RNA. Nat. Methods. 2019; 16:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M.S., Dai Q., Di Segni A., Salmon-Divon M., Clark W.C. et al.. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. 2016; 530:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Davalos V., Blanco S., Esteller M.. SnapShot: messenger RNA modifications. Cell. 2018; 174:498–498. [DOI] [PubMed] [Google Scholar]
- 58. Liu Q., Fang L., Yu G., Wang D., Xiao C.L., Wang K.. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 2019; 10:2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated fastq data in this study were deposited in the SRA database (accession number SRP166020). ELIGOS is available at https://gitlab.com/piroonj/eligos2.