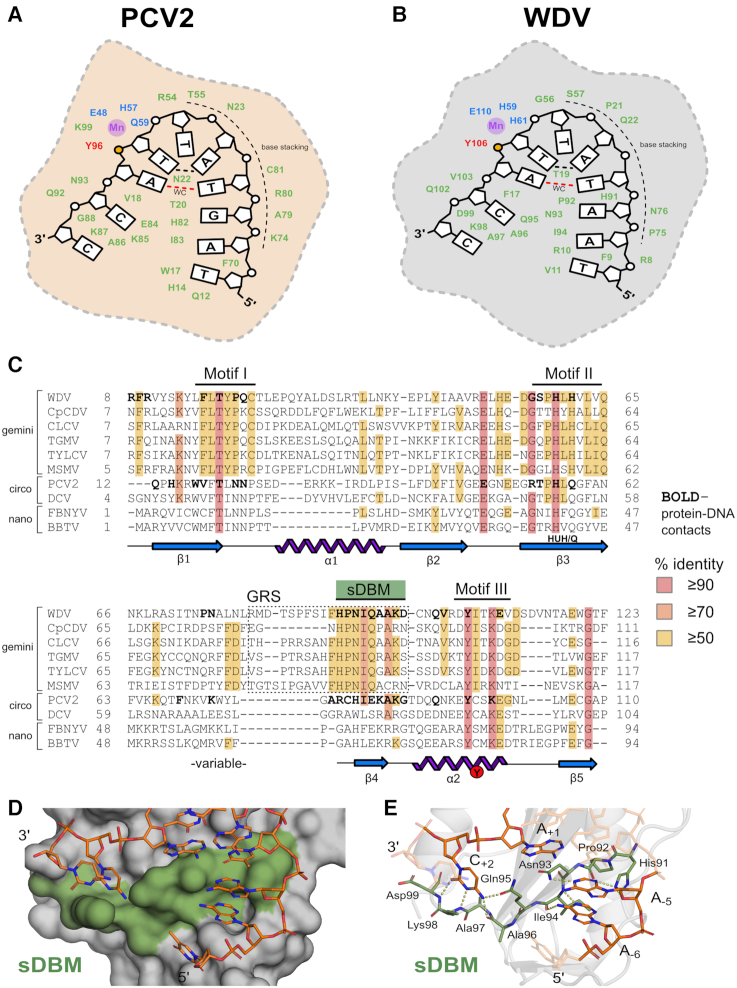
Figure 2.
Cartoon depiction and structural alignment of specific Rep protein–DNA interactions: (A) PCV2 and (B) WDV Rep structures depicted as 2D cartoons with relative positions of residues (green) involved in binding ‘U-shaped’ ssDNA within 4 Å. The catalytic tyrosine 106 is indicated in red with the adjacent phosphate in yellow, the ion coordinating triad is indicated in blue, and the 2+ ion in purple. The single Watson–Crick (WC) base pair is indicated as a dashed red line, and the ssDNA intramolecular hydrogen bond is indicated as a black dashed line. (C) Structural alignment of Reps using PROMALS3D including available PCV2 (PDB: 6WDZ), WDV (PDB: 6WE0), TYLCV (1L2M) and FBNYV (6H8O) structures as templates with conserved residues highlighted - high or absolute conservation (≥90%) indicated in red; moderately conserved (≥70%) indicated in orange; low conservation (≥50%) indicated in yellow; and no conservation (<50%) indicated in white. Amino- and carboxy- terminal ends are trimmed to reflect only structured domains in crystal structures. Bolded residues indicate contacts within 4 Å of DNA 10-mers complexed with PCV2Y96F (PDB: 6WDZ) and WDVY106F (PDB: 6WE0). Conserved Rep Motifs I/II/III are shown as well as the GRS motif within the dashed box for geminivirus Reps. The sDBM we have defined in this study is labelled and highlighted in green. The conserved secondary structural elements making up the core nickase domain (β1–5 and α1–2) below the alignment sequences are shown as 2D cartoons with labeled HUH/Q motif and catalytic tyrosine. (D) Surface representation of WDV with sDBM highlighted in green bound to the 10-mer as sticks. (E) Major polar interactions between WDV sDBM residues (green sticks) and bases of 10-mer (orange sticks) are shown as yellow dashes.