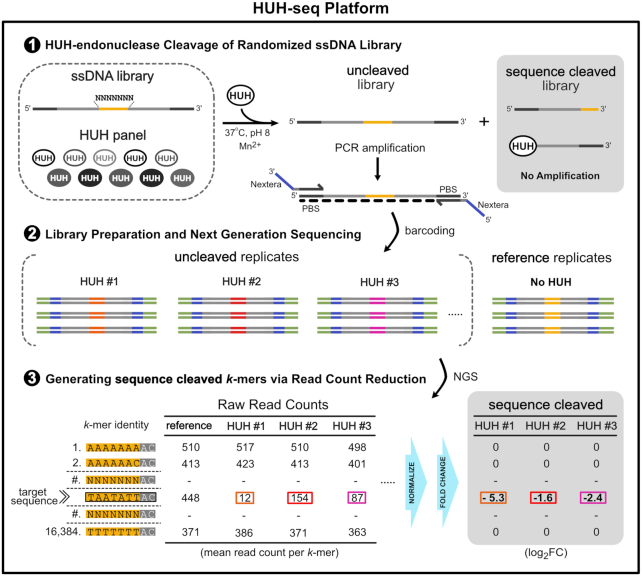
Figure 4.
HUH-seq cleavage assay schematic for determining Rep sequence specificity. Schematic describing HUH-seq: an NGS-based approach for quantifying ssDNA specificity profiles of Reps. A synthetic ssDNA library containing seven random bases (4 bases ∧ 7 positions = 16 384 unique kmers) (yellow) flanked by constant regions (gray) and primer binding sites (PBS) (dark gray) are reacted with a panel of Reps, or no enzyme as a reference, in replicate, generating a two part pool containing the ‘uncleaved’ library and the ‘sequence cleaved’ library for each reaction. In a single PCR step, the antisense strand for the ‘uncleaved’ pool is generated, amplified and Nextera adapters (purple) are added with primer overhangs; the ‘sequence cleaved’ library is not amplified due to physical separation of the PBS’s. Each set of amplicons is then barcoded with standard i7/i5 Illumina indexing sequences (green) and pooled for a single next generation sequencing run. A custom R-based analysis script generates read counts for all k-mers in each set of replicates, then normalizes based on total read count, and quantifies k-mer cleavage extent of each Rep in the panel based on fold change and percent reduction.