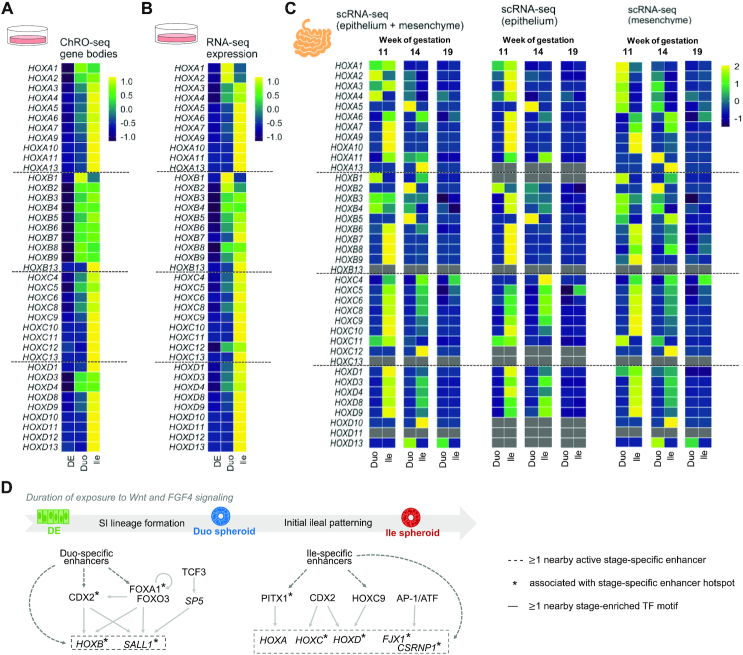
Figure 7.
The HOX cluster dynamics observed in the directed differentiation model of SI is present in the early developmental stages of primary human fetal SI tissue. (A) Heatmap showing the changing transcriptional pattern of HOX genes (ChRO-seq) across the stages of DE, Duo spheroids and Ile spheroids in vitro. (B) Heatmap showing changing steady-state expression pattern of HOX genes (RNA-seq) across stages of DE, Duo spheroids and Ile spheroid in vitro. (C) Heatmap showing changing expression pattern of HOX genes (scRNA-seq) in primary human fetal Duo and Ile tissues at week 11, 14 and 19 of gestation. The expressional level of each HOX gene is the average from the fraction that has both epithelial and mesenchymal cell types (left), mesenchymal cell only (middle), or epithelial cell only (right). (D) Proposed model of transcriptional programming in SI lineage formation and ileal regional patterning during human SI development. The genes activated in the relevant stages and the types of regulatory relationships identified through ChRO-seq analyses are indicated. ChRO-seq study: DE, n = 4; Duo spheroid (Duo), n = 3; Ile spheroid (Ile), n = 3. RNA-seq study: DE, n = 3; Duo, n = 6; Ile, n = 4. scRNA-seq data: matched Duo and Ile tissues from one human fetal subject per gestational time point.