Figure 1.
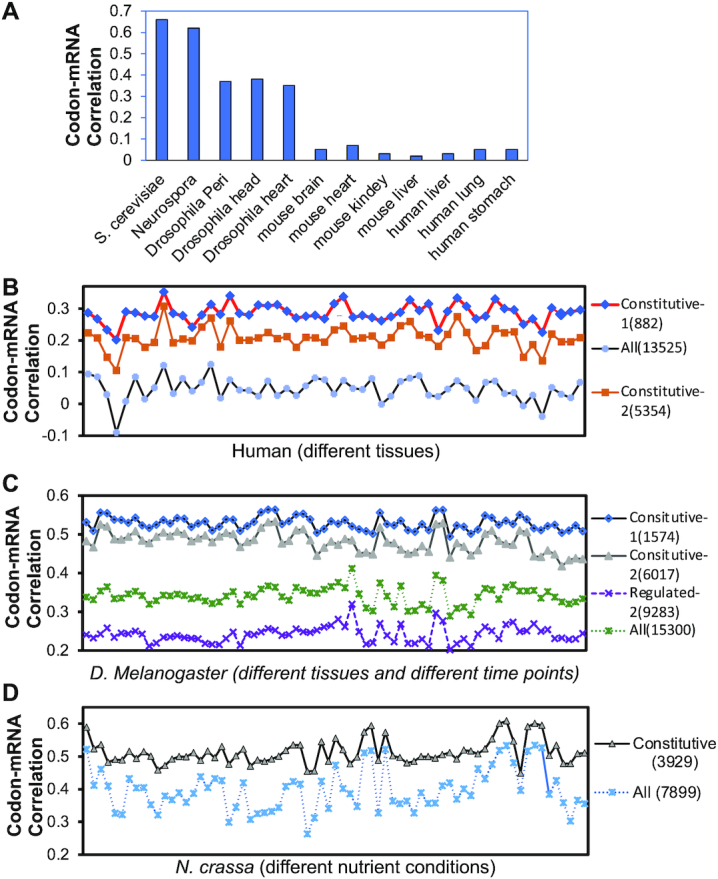
Differential genome-wide correlations between codon usage and mRNA levels between constitutive and regulated genes from fungi to human cells and tissues. (A) Pearson correlation coefficients between genome-wide gene codon usage and mRNA levels in the indicated organisms and tissues. (B–D) Pearson correlation coefficients between genome-wide gene codon usage and mRNA levels in B) various human tissue samples, C) Drosophila tissue samples collected at various time points, and D) Neurospora samples collected in different nutrient conditions. tAI was used to determine gene codon usage of human and Drosophila genes (B, C) and CBI was used to determine codon usage of N. crassa genes (D). The number of genes per category are indicated in parentheses. Each symbol indicates one tissue, one time points or one nutrient condition. The RNA-seq samples in B–D are listed in Supplementary Table S2.
