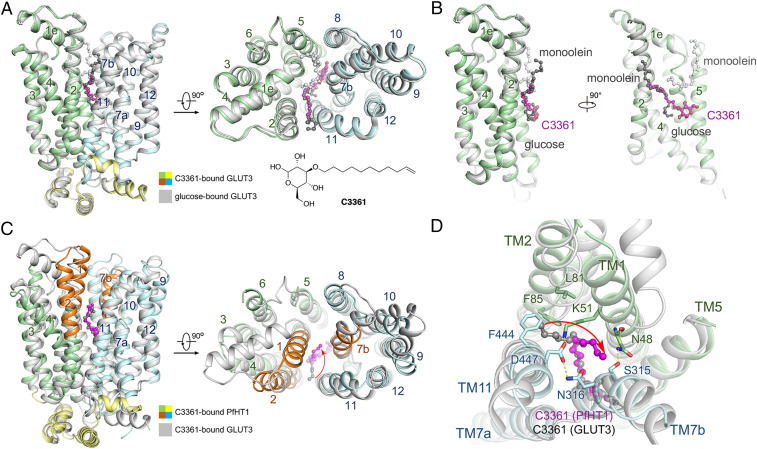
Fig. 2.
Crystal structure of hGLUT3 bound to C3361 in an outward-occluded conformation. (A) Superimposed structures of glucose–hGLUT3 complex (gray, PDB ID code 4ZW9) and C3361–hGLUT3 complex (domains colored) presented in both side and intracellular views of overall structures. The protein structures are shown in cartoon representation, and the ligands are shown in ball-and-stick representation. The amino-terminal, carboxyl-terminal, and intracellular helical domains of C3361-bound hGLUT3 are colored in pale green, pale cyan, and yellow, respectively. (B) The coordination of the sugar moiety of C3361 is nearly identical to that of d-glucose, and the tail of the C3361 occupied the position where a monoolein molecule located in the glucose-bound hGLUT3 structure. (C) Superimposed structures of C3361–hGLUT3 complex (gray) and C3361–PfHT1 complex (domains colored, PDB ID code 6M2L) presented in both side and intracellular views of overall structures. (D) C3361 demonstrated distinct binding modes when complexed with hGLUT3 or PfHT1; in particular, its tails pointed toward different directions.