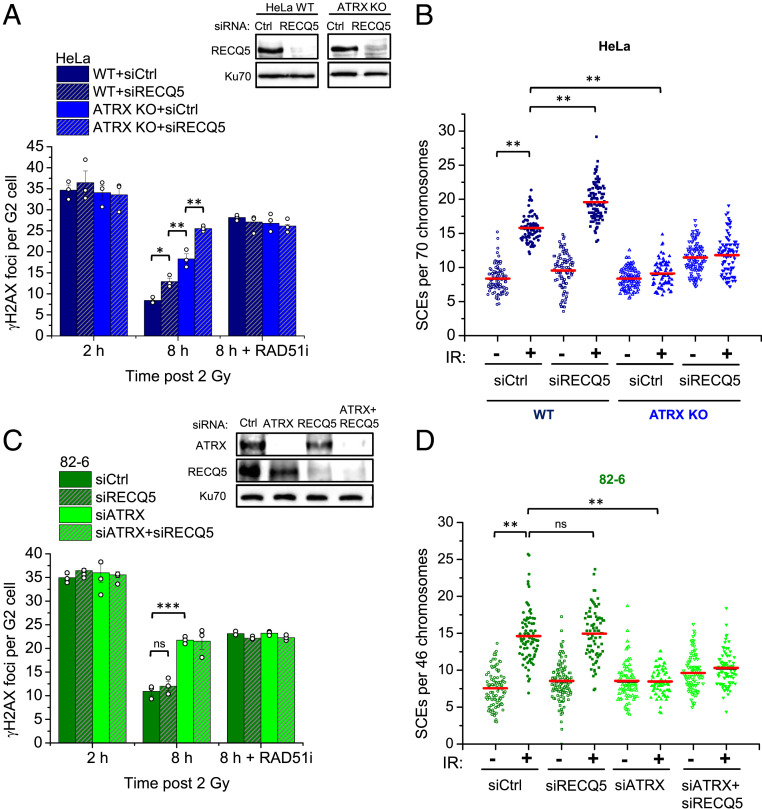
Fig. 3.
HR subpathway usage in ATRX-proficient cancer and normal cells. (A) HeLa WT and ATRX KO cells were transfected with siCtrl or siRECQ5 and treated with RAD51i prior to IR and throughout repair incubation. γH2AX foci were enumerated in EdU-negative G2 cells. Spontaneous foci (three to four) were subtracted. Knockdown was confirmed by immunoblotting. Data for siCtrl were extracted from supplemental appendix, figure S3A in ref. 26 as they were part of the same experiment with siRECQ5. (B) HeLa WT and ATRX KO cells were transfected with siCtrl or siRECQ5 and incubated with BrdU for 48 h. Cells were then irradiated with 2 Gy, collected after 8 h, and processed to obtain mitotic spreads. SCEs per spread were quantified and normalized to 70 chromosomes. (C) 82-6 cells were transfected with siCtrl, siRECQ5, and/or siATRX and treated with RAD51i prior to IR and throughout repair incubation. γH2AX foci were enumerated in EdU-negative G2 cells. Spontaneous foci (one to two) were subtracted. Knockdown was confirmed by immunoblotting. (D) 82-6 cells were transfected with siCtrl, siRECQ5, and/or siATRX and incubated with BrdU for 48 h. Cells were then irradiated with 2 Gy, collected after 12 h, and processed to obtain mitotic spreads. SCEs per spread were quantified and normalized to 46 chromosomes. Foci data show mean ± SEM (n = 3). Results from individual experiments, each derived from 40 cells, are indicated. Individual SCE data are shown and red horizontal lines indicate the mean; 70 to 120 spreads and >4,000 chromosomes per condition were analyzed from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant (two-tailed t test).