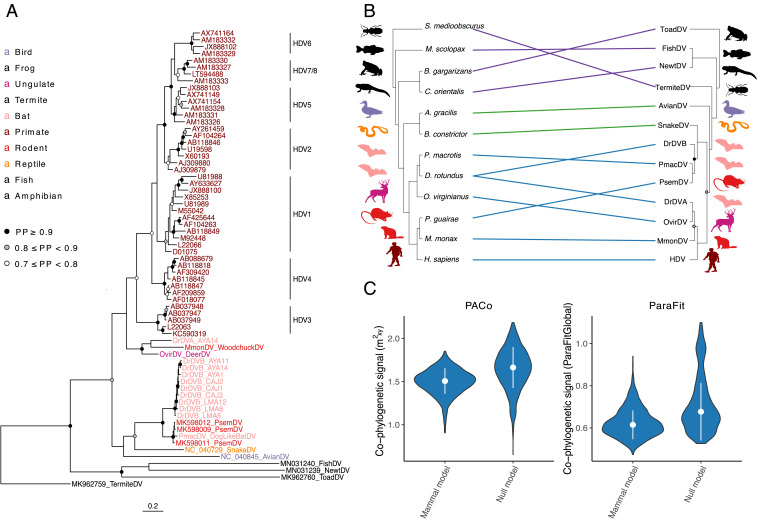
Fig. 2.
The evolutionary history of deltaviruses reveals host shifts among mammals. (A) Bayesian phylogeny of a 192-aa alignment of the DAg. Ingroup taxa, including mammal, snake, and avian deltaviruses, are colored by order; other HDV-like taxa are shown in black. (B) Cophylogeny depicting connections between the consensus deltavirus phylogeny from StarBeast and the host tree (56). Links are colored according to subsets of data used in cophylogenetic analyses; all taxa (purple + green + blue), ingroup (green + blue), or mammal (blue). Host taxa are Schedorhinotermes medioobscurus, Macroramphosus scolopax, Bufo gargarizans, Cynops orientalis, Anas gracilis, Boa constrictor, Peropteryx macrotis, Desmodus rotundus, Odocoileus virginianus, Proechimys guirae, Marmota monax, and Homo sapiens. (C) An absence of phylogenetic dependence of the mammalian deltavirus phylogeny on the host phylogeny. Violin plots show distributions of test statistics from two cophylogenetic approaches across 1,000 posterior trees relative to null models, along with medians and SDs. For PACo, higher values would indicate greater phylogenetic dependence; for ParaFit, lower values would indicate greater phylogenetic dependence. Both approaches rejected a global model of cospeciation (P > 0.05).