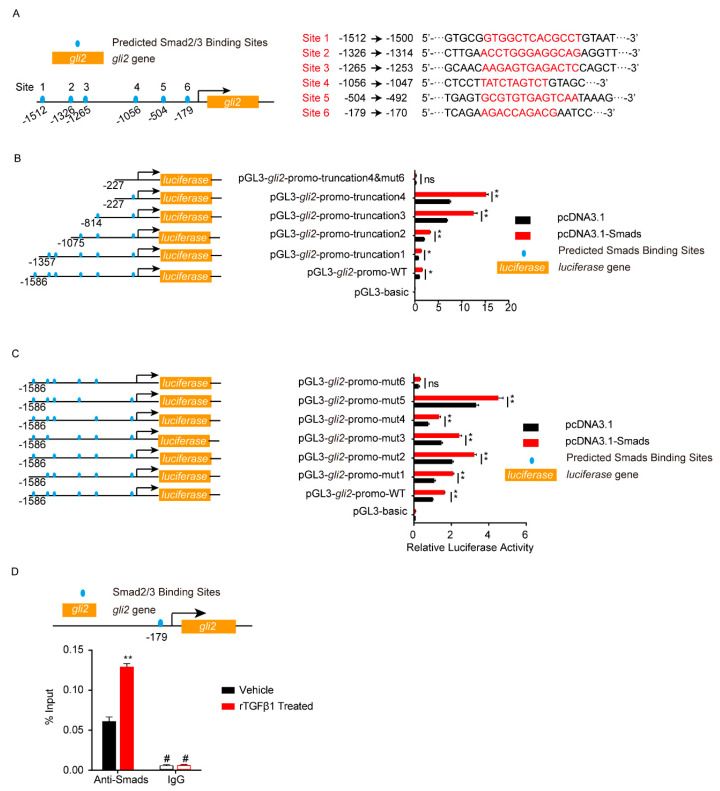
Figure 5.
Analysis of Smad2/3 binding sites on gli2 promotor by dual-luciferase reporter assays. (A) Schematic of the 6 predicted Smad2/3 binding sites on gli2 promotor (left) and their binding sequences accordingly (right). The binding sites were located at −1512 to −1500 (site 1), −1326 to −1314 (site 2), −1265 to −1253 (site 3), −1056 to −1047 (site 4), −504 to −492 (site 5), and −179 to −170 (site 6) of the gli2 promotor region. (B–C) The gli2 luciferase activity was tested by applying a series of truncations (B) and site-targeted mutations (C) to the gli2 promoter, along with pcDNA3.1-Smad2 and pcDNA3.1-Smad3 and pRL-TK plasmids. The specific constructs used in the truncation assays (B) included the pGL3-basic vector, pGL3-gli2-promo-WT (containing promotor region from −1568 to +35), pGL3-gli2-promo-truncation1 (from −1357 to +35), pGL3-gli2-promo-truncation2 (from −1075 to +35), pGL3-gli2-promo-truncation3 (from −814 to +35), pGL3-gli2-promo-truncation4 (from −227 to +35), and pGL3-gli2-promo-truncation4&mut6 (from −227 to +35 which lacks site 6). The specific constructs used in the site-mutation assays (C) included the pGL3-basic vector, pGL3-gli2-promo-WT (containing all 6 sites), pGL3-gli2-promo-mut1 (lack of site 1), pGL3-gli2-promo-mut2 (lack of site 2), pGL3-gli2-promo-mut3 (lack of site 3), pGL3-gli2-promo-mut4 (lack of site 4), pGL3-gli2-promo mut5 (lack of site 5), and pGL3-gli2-promo-mut6 (lack of site 6). The luciferase activities were determined, and are presented as the ratios of firefly luciferase activity and renilla luciferase activity. The assays were performed with 3 replicates, and data are presented as mean ± SEM. * p < 0.05, ** p < 0.01. ns, no significance. (D) ChIP-qPCR validation of the Smad2/3 binding to the gli2 promotor around −179 in hBMECs treated by TGFβ1 (50 ng/mL). ChIP was performed with anti-Smad2/3 antibody and rabbit IgG was employed as the negative control. # indicates lower than the detection limits. ** p < 0.01.