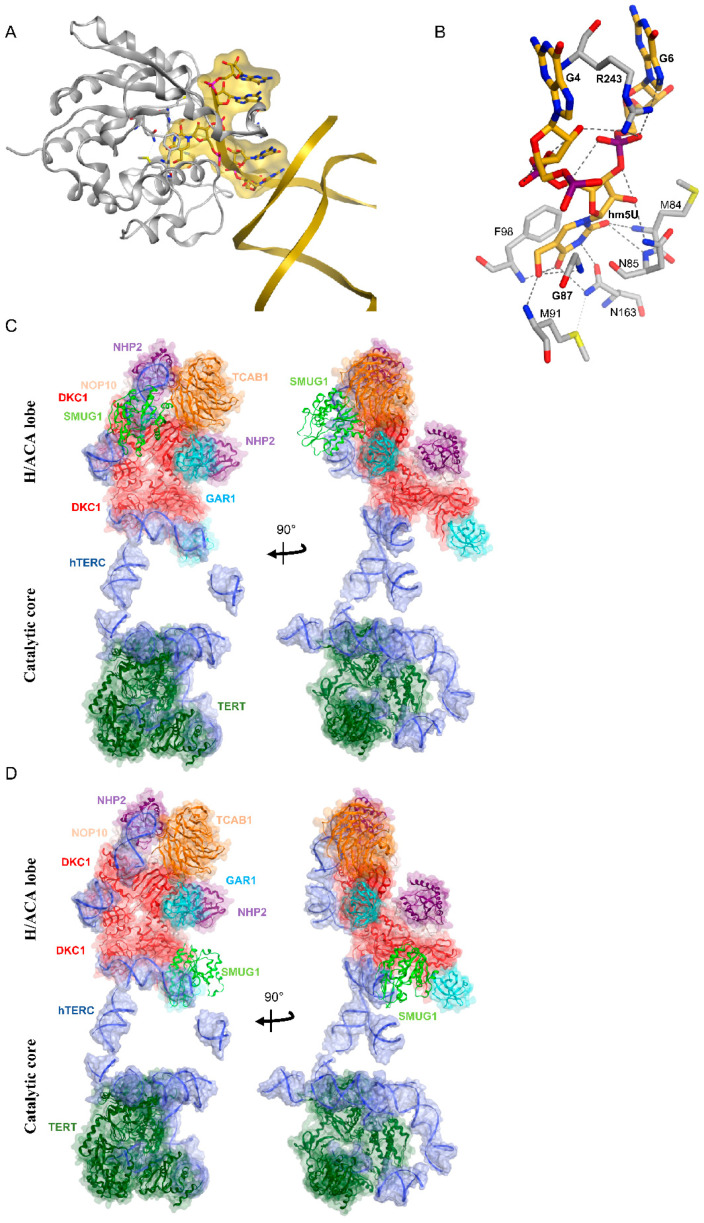
Figure 6.
Computationally predicted binding pose of hSMUG1 to TERC and the human telomerase complex. (A) Homology model of hSMUG1 (shown in silver ribbons) bound to the fish TERC structure (shown in gold ribbons), which is computationally modified to mimic hTERC at the binding interface. The residues of hSMUG1 that interact with the flipped hm5U are shown in sticks with C atoms in silver. The flipped hm5U as well as the two nucleotides neighboring it on each side in TERC are depicted in sticks with C atoms in gold. Both for the residues and the nucleotides, N atoms are in blue, O atoms are in red, S atoms are in yellow, and P atoms are in magenta. The terminal 5 nucleotides of TERC are additionally highlighted depicting their molecular surface in gold. (B) Close-up view of panel A depicting the interaction of hSMUG1 active site with the flipped hm5U at position 220 and its neighboring nucleotides. Everything except the interacting residues is hidden to provide a clear view. The color code is the same as in panel A, except here P atoms are depicted in purple. Interactions are displayed as dotted lines. (C,D) Superimposing SMUG1 on the pdb structure of human telomerase complex cryo-EM. Our prior experimentally validated SMUG1–DKC1 complex structure is superimposed onto each of the two dyskerin (DKC1) units found in human telomerase. In both cases, a significant steric clash of SMUG1 with the rest of the human telomerase is predicted.