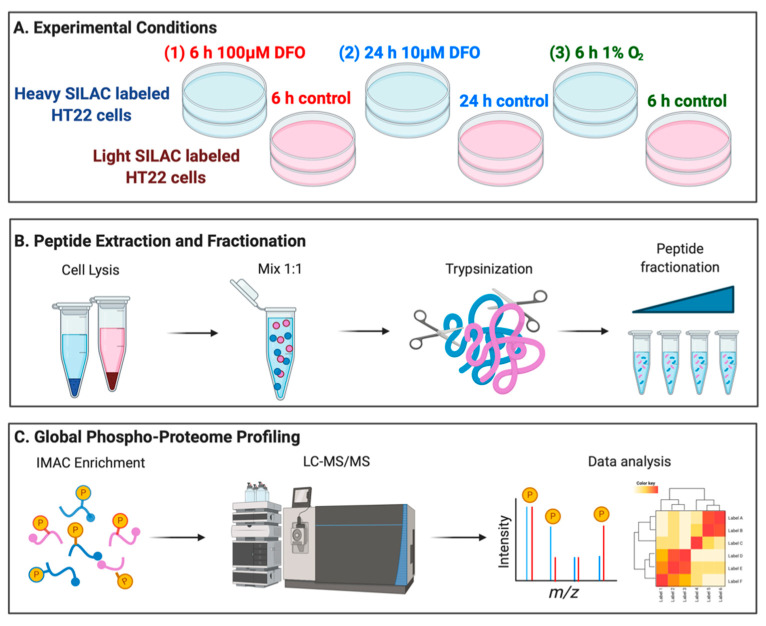
Figure 2.
Experimental design and phosphoproteomic workflow for comprehensive analysis of phosphorylation proteome. (A) Outline of experimental workflow using Stable Isotopic Labeling of Amino acids in Cell culture (SILAC)-labeled HT22 cells, (B) generating peptides for quantification, and (C) enrichment of phospho-sites, mass spectrometry acquisition, and data analysis.