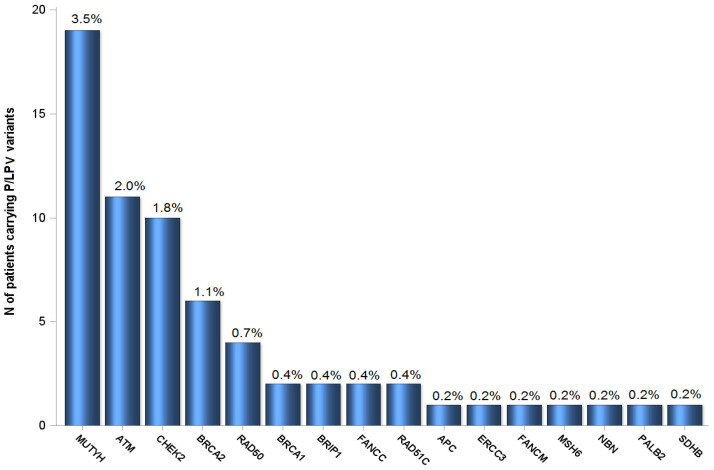
Figure 1.
Prevalence of P/LPVs in 549 patients of the study. Single nucleotide variants (SNV), insertions/deletions (Indels) and copy number variant (CNV) detection was performed for the following genes: APC, ATM, BAP1, BARD1, BMPR1A, BRCA1, BRCA2, BRIP1, CDH1, CDK4, CDKN2A, (CDKN2Ap16(INK4A), CDKN2Ap14(ARF)), CHEK2, DDB2, DICER1, EPCAM, ERCC1, ERCC2, ERCC3, ERCC4, ERCC5, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, GREM1, HOXB13, MEN1, MLH1, MRE11, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, POLD1, POLE, POLH, PTEN, RAD50, RAD51C, RAD51D, RB1, RET, SDHAF2, SDHB, SDHC, SDHD, SLX4, SMAD4, SMARCA4, STK11, TP53, VHL, XPA, XPC. Overall, 62 patients (11.3%) carried ≥1 P/LPV in at least 1 of 16 genes. Three patients carried two P/LPVs; two carried P/LPVs in CHEK2 and MUTYH and one in BRCA1 and MUTYH. The rest of the patients (59) had one P/LPV detected. The percentage of patients carrying a P/LPV in each gene is shown on top of each bar.