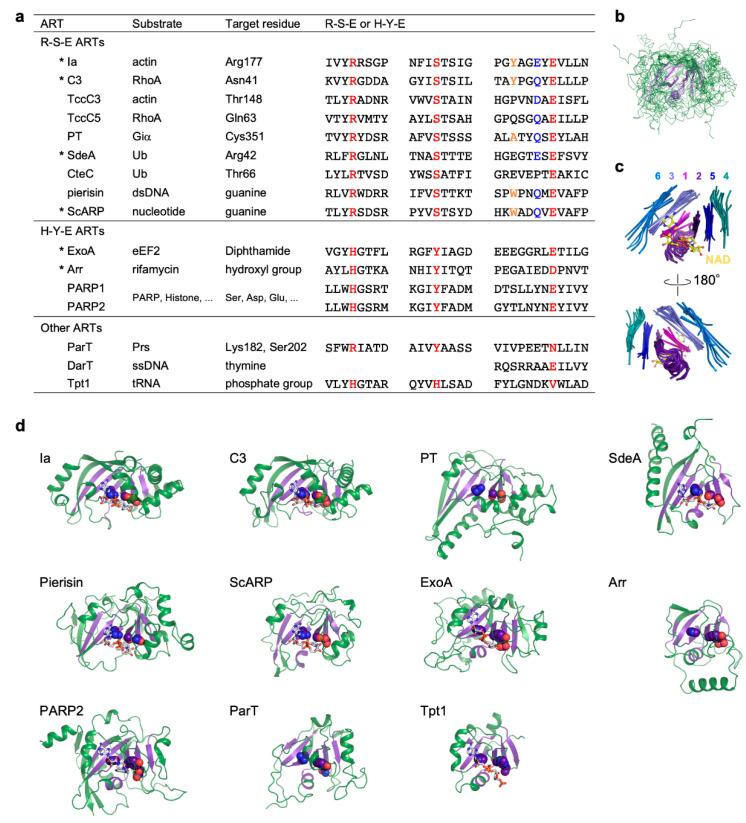
Figure 1.
Comparison of R-S-E class, H-Y-E class, and other ADP-ribosyltransferases. (a) Substrate, target residue, and conserved motifs. Conserved R-S-E and H-Y-E residues are shown in red. In the R-S-E motif, the third aromatic residue and the sixth glutamate or glutamine residue in the ADP-ribosylating toxin turn-turn (ARTT) loop (X-X-ϕ-X-X-E/Q-X-E) are shown in orange and blue, respectively. ADP-ribosyltransferases (ARTs) with available substrate-complexed structures are indicated with asterisks. (b) Superimposed structures of the ART domains of the 11 ARTs are shown in (d). The structure of the ART core, including six strands and the helix, are shown in violet. (c) Close-up views of ART core structures. Individual strands are shown in different colors and are numbered from the N-terminus to the C-terminus. NAD bound to Ia is shown as a ball-and-stick model. (d) Structures of the ART domain. ART core structures are shown in violet. R-S-E and H-Y-E residues are shown as sphere models. NAD or NAD analogs are shown as stick models. Protein Data Bank (PDB) IDs: Ia, 4h03; C3, 4xsh; PT, 1bcp; SdeA, 5yij; Pierisin, 5h6j; ScARP, 5zj5; ExoA, 2zit; Arr, 2hw2; PARP2, 6tx3; ParT, 6d0h; Tpt1, 6e3a.