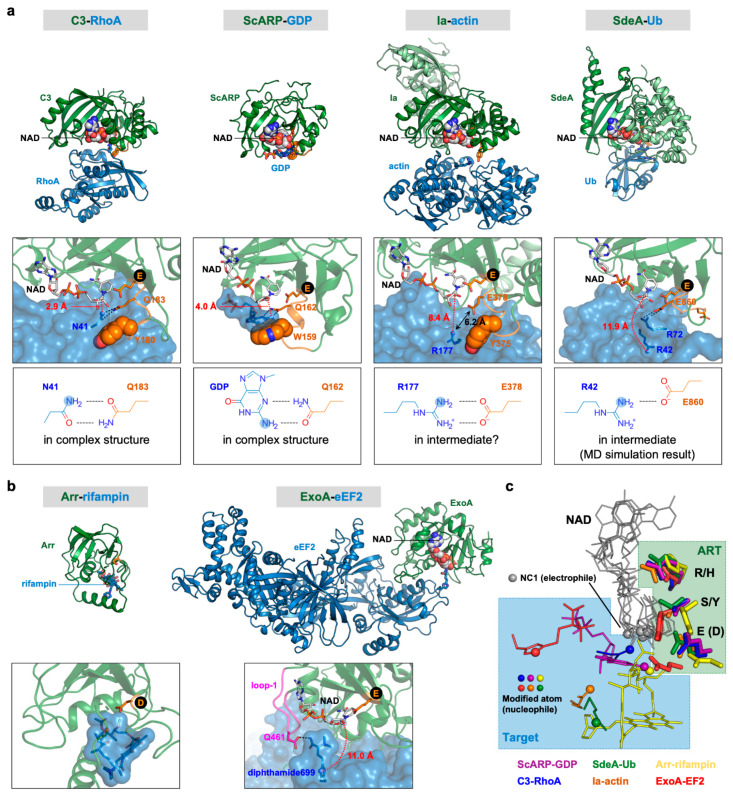
Figure 2.
Comparison of complex structures of ARTs and their substrates. (a) R-S-E class ARTs. Top, overall structures; middle, close-up views of active sites; bottom, interactions between the target and the sixth amino acid of the ARTT loop. (b) H-Y-E class ARTs. Top, overall structures; bottom, close-up views of active sites. The N-terminal domain of Ia and the α-helical domain of SdeA are shown in light green color. The ARTT loops of R-S-E class ARTs are shown in orange. The NC1 of NAD (electrophile) and the modified atom (nucleophile) are shown as sphere models. Distances between two atoms involved in forming the new bond, namely the NC1 of NAD (electrophile) and the modified atom (nucleophile), are shown in red. Black circles indicate the third catalytic glutamate residues of the R-S-E and H-Y-E motifs. (c) Positional relationships between nucleophiles and NADs. Superimposition of six complex structures are shown in (a,b). R-S-E and H-Y-E residues are shown as stick models. NADs and targets are shown as lines. NC1 atoms of NADs (electrophiles) and modified atoms (nucleophile) are shown as sphere models. PDB IDs: Ia–actin, 4h03; C3–RhoA, 4xsh; SdeA–Ub, 5yij; ScARP–GDP, 5zj5; ExoA–eEF2, 2zit; Arr–rifampin, 2hw2.