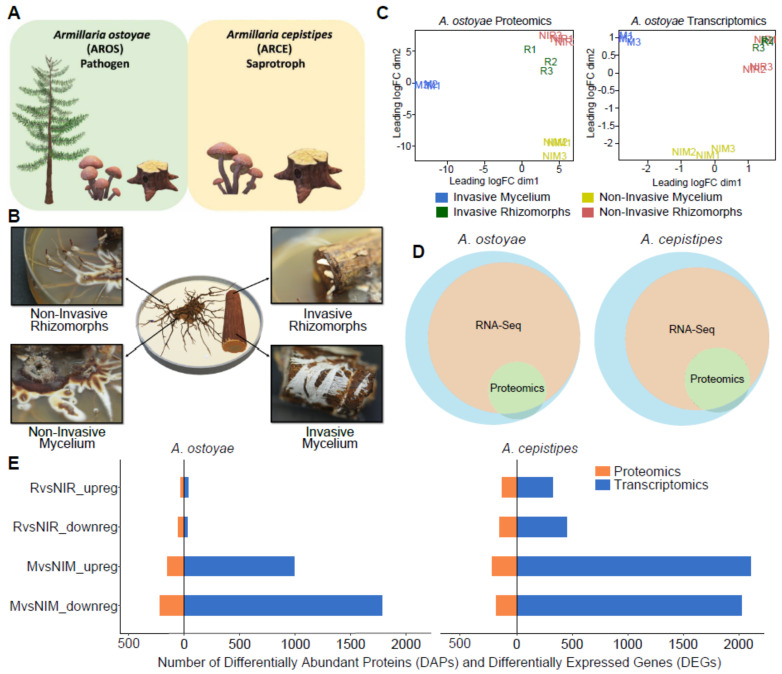
Figure 1.
Overview of the experimental approach for root decay studies. (A) Representation of Armillaria ostoyae (pathogenic) and A. cepistipes (saprotroph) used in this study, (B) The four tissue types sampled for transcriptomics and proteomics analysis, viz., invasive mycelium (growing beneath the outer layer of root), invasive rhizomorphs (emerging out of the roots), non-invasive mycelium and non-invasive rhizomorphs (growing in absence of root). A. ostoyae is shown as an example, (C) Multidimensional scaling of three biological replicates from each of the tissue types in A. ostoyae for proteomics (left) and transcriptomics (right), (D) Proportion of transcripts and proteins detected in the two -omics analysis. The blue circle represents the whole proteome, orange depicts the transcripts detected in the RNASeq, and green represents the proteins detected in the proteomics analyses. (E) The number of differentially expressed genes (blue) and differentially abundant proteins (orange) detected in the two species.