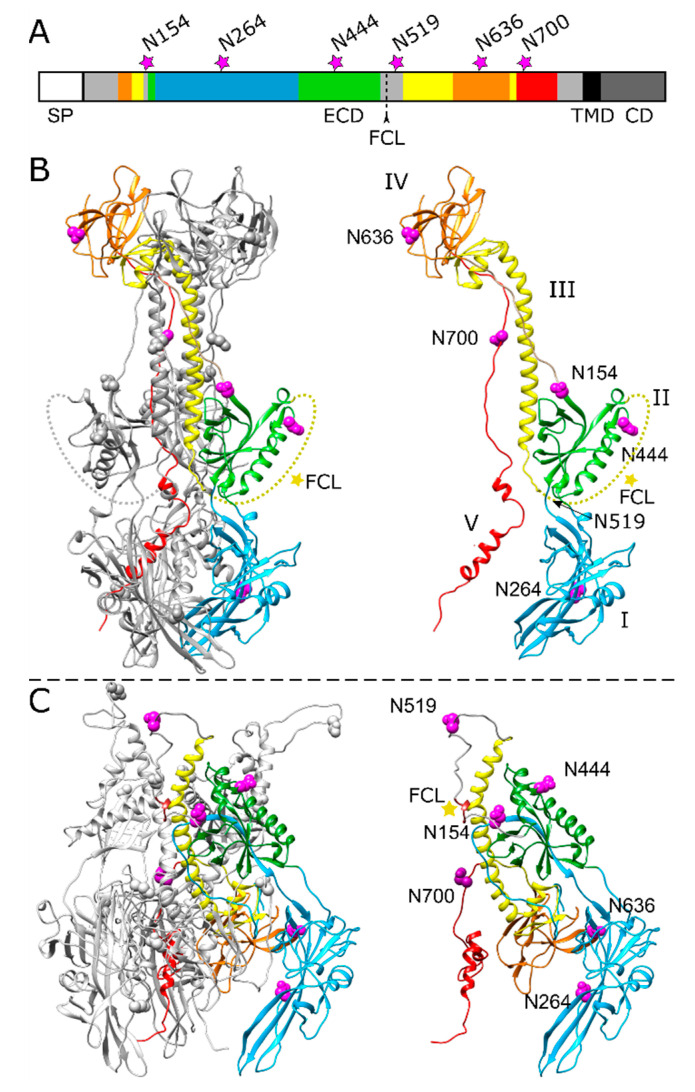
Figure 1.
Position of potential N-linked glycosylation sites in the PrV gB ectodomain. (A) Schematic diagram of PrV gB with the signal peptide (SP), ectodomain (ECD), transmembrane domain (TMD) and a cytoplasmic domain (CD) indicated. The five domains (DI-V) forming the ectodomain are colored in blue (DI), green (DII), yellow (DIII), orange (DIV) and red (DV) according to the ribbon diagrams in (B,C), and regions not resolved in the post-fusion structure are depicted in grey. Position of the furin-cleavage site (FCL) and the potential asparagine (N)-linked glycosylation sites N154, N264, N444, N519, N636 and N700 are indicated. (B) Ribbon diagram of the PrV gB post-fusion trimer (PDB ID: 6ESC) [18], (left panel) and the monomer (right panel) is shown. Domains I–V of one protomer are highlighted in different colors, and the predicted glycosylation sites are indicated by purple spheres. N519 lies within a flexible region (dotted yellow line) that was not solved in the crystal structure and is marked by an arrow. The yellow star indicates the furin-cleavage site. (C) Model of pre-fusion PrV gB trimer (left panel) was generated using the protein structure homology-modeling server SWISS-MODEL [26] and the pre-fusion structure of HSV-1 gB as template [24]. Domains I–V of one protomer (right panel) are colored as in (A,B), and the predicted glycosylation sites are shown as purple spheres, and the furin-cleavage site is marked by a yellow star. The structure images were generated using UCSF Chimera (version 1.13.1) [27].