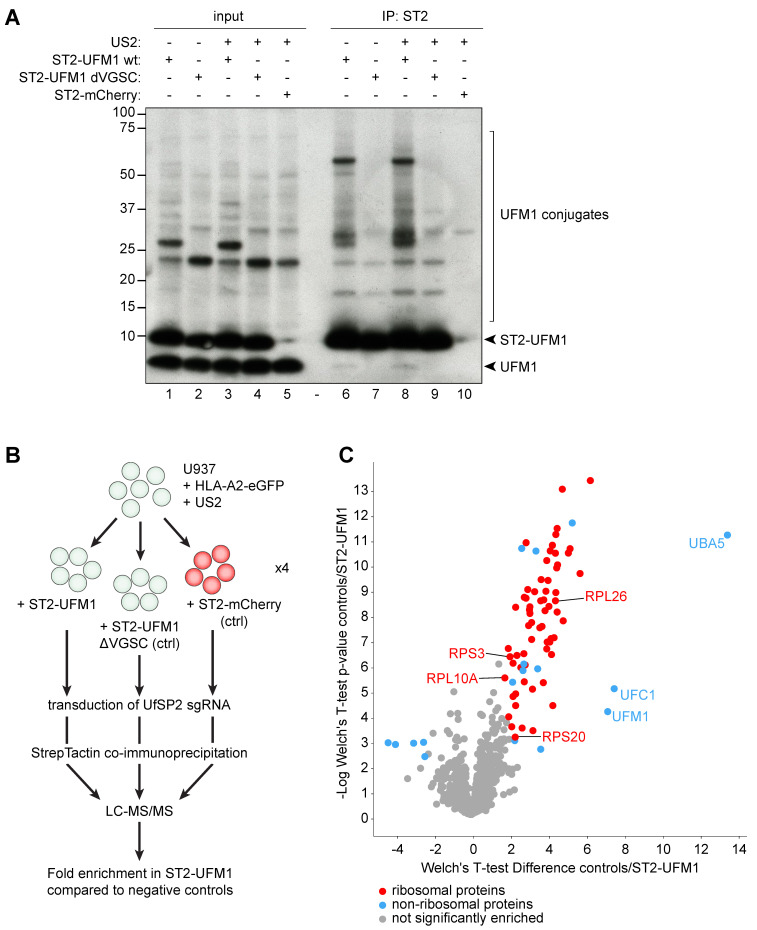
Figure 5.
Mass spectrometry identifies ribosomal UFMylation in the presence of US2. (A) U937 cells expressing HLA-A2-eGFP in the presence of absence of US2 were transduced with StrepII-tagged UFM1 wildtype, a StrepII-tagged ΔVGSC mutant of UFM1, or StrepII-tagged mCherry. A sgRNA targeting UfSP2 was subsequently introduced in all cell lines. StrepTactin co-immunoprecipitations were performed in 1% LMNG to pull down UFM1 or control substrates. Immunoblotting was performed with an antibody staining against UFM1. (B) Schematic set-up of the cell lines and workflow used for mass spectrometry. (C) Volcano plot showing the proteins identified by mass spectrometry upon StrepII-UFM1 immunoprecipitation. The enrichment per protein was calculated compared to the negative control cell lines. Significantly enriched (top right) or depleted (left) hits are shown in color, while the gray dots show proteins that are not significantly enriched. Red hits represent ribosomal proteins, while all other hits are shown in blue. A complete list of names and functions of the proteins that were significantly enriched or depleted is presented in Supplementary information 2.