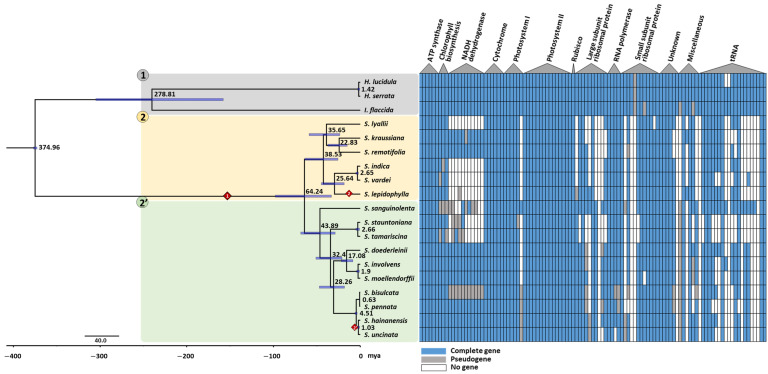
Figure 3.
Phylogenetic analysis of the 19 lycophyte species examined in this study. The coding sequence (CDS) of 45 shared genes were used in the phylogenetic analysis with BEAST software. The calibration node was set to 375 mya for the split of Isoetes and Selaginella species. Left, phylogenetic tree. The gray box labeled 1 highlights non-Selaginella species. Lineages 2 and 2′ in the Selaginella genus are highlighted with yellow and green boxes, respectively. The red diamond labeled 1 represents the first inversion event, which differentiates non-Selaginella species from Selaginella species. Red diamonds labeled 2 and 2′ represent the second inversion events, which occurred independently in each lineage and converted the direct repeat structures back into inverted repeats. Blue bars on the branch of the phylogenetic tree indicate height 95% highest posterior density (HPD) and numbers near the nodes represent median values. Right, corresponding gene contents for each species. Blue boxes indicate genes that are present, gray boxes indicate pseudogenes, and white boxes indicate missing genes.