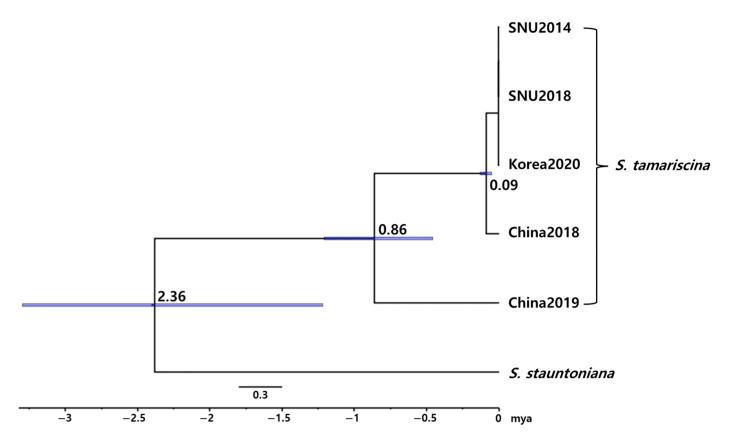
Figure A1.
Phylogenetic tree of five Selaginella tamariscina plastid genome sequences with Selaginella stauntoniana as the outgroup. Phylogenetic analysis was performed with protein coding sequences that all six plastome sequences share with BEAST v2.5.2 software. The HYK substitution model and a gamma category count of 4 were used in the Gamma Site Model. For this analysis, a strict clock with a clock rate of 1, as well as the coalescent constant population tree prior, were applied. 2.66 mya, representing the split between S. tamariscina and S. stauntoniana derived from the phylogenetic analysis in this study (Figure 3), was used as the secondary calibration value with a normal distribution. The five S. tamariscina species were grouped into one monophyletic group, and the chain length was set to 1 M, sampling every 1000 chains. All other values were left at default parameters. Results were checked for the effective sample size (ESS) via Tracer v1.7.1, and the maximum clade credibility tree was drawn by TreeAnnotator with a burnin of 15%. Visualization of the final tree was conducted with FigTree v1.4.4. Blue bars show node bars representing the height 95% HPD, and numbers near the nodes show the median values.