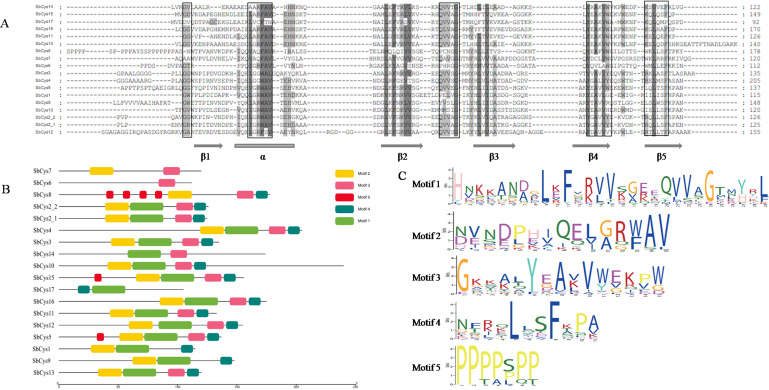
Figure 3. The amino acid alignment and conserved motifs distribution of SbCys.
(A) The locations of the secondary structures (α-helix and β-sheets) were included. The main cystatin conserved motifs are in black boxes. The strong and weak conservative changes in amino acids are marked by dark gray and light gray font, respectively. (B) The motifs were identified by MEME. Each motif was represented by one color box. (C) Conserved protein motif 1 (QxVxG), motif 2 ( LARFAV and G-residue), motif 3 (W-residue), motif 4 ([RK]xLxxF), and motif 5(P-residue) presented in the variable region of cystatin genes.