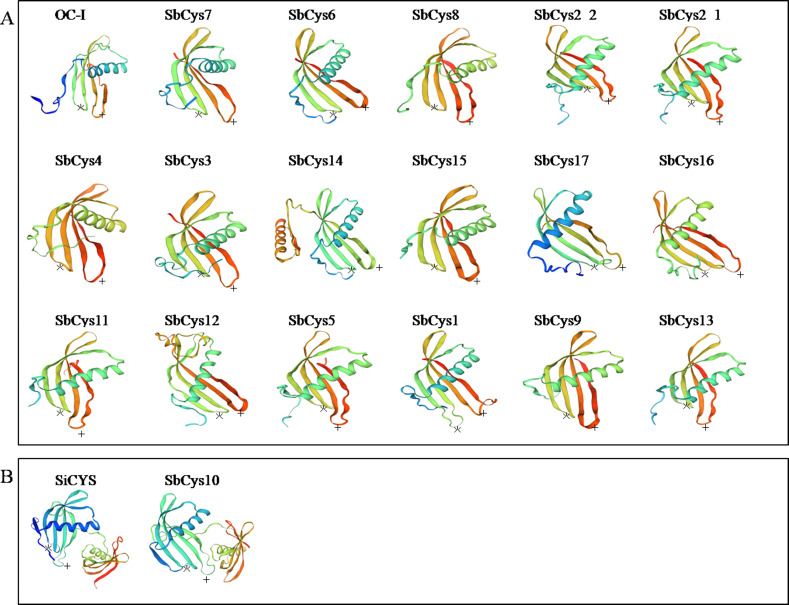
Figure 4. The three-dimensional structure prediction of Sorghum cystatins.
(A) The three-dimensional structures of SbCys proteins were predicted using the automated SWISS-MODEL program with OC-I as a template. (B) The three-dimensional structure of SbCys10 was predicted using the automated SWISS-MODEL program with SiCYS as a template. Two important motifs involved in the interaction with the target enzymes are indicated: the reactive site (asterisks) and W residue (crosses).