Abstract
Motivation
Oxford Nanopore technologies (ONT) add miniaturization and real time to high-throughput sequencing. All available software for ONT data analytics run on cloud/clusters or personal computers. Instead, a linchpin to true portability is software that works on mobile devices of internet connections. Smartphones’ and tablets’ chipset/memory/operating systems differ from desktop computers, but software can be recompiled. We sought to understand how portable current ONT analysis methods are.
Results
Several tools, from base-calling to genome assembly, were ported and benchmarked on an Android smartphone. Out of 23 programs, 11 succeeded. Recompilation failures included lack of standard headers and unsupported instruction sets. Only DSK, BCALM2 and Kraken were able to process files up to 16 GB, with linearly scaling CPU-times. However, peak CPU temperatures were high. In conclusion, the portability scenario is not favorable. Given the fast market growth, attention of developers to ARM chipsets and Android/iOS is warranted, as well as initiatives to implement mobile-specific libraries.
Availability and implementation
The source code is freely available at: https://github.com/marco-oliva/portable-nanopore-analytics.
1 Introduction
Third-generation DNA/RNA sequencing added miniaturization and portability to cost-effective, high-throughput data yield of the second-generation sequencers, e.g. Illumina or Ion Torrent (Jain et al., 2016; Low and Tammi, 2017). For example, Oxford Nanopore technologies (ONT) include the MinION, which weighs 90 g and measures 10×3×2 cm. MinION’s throughput ranges from a few to 20 GB, sequence read length ranges from thousands to hundred thousand of nucleotide bases, and many sequencing kits complete in <1 h (Ma et al., 2017). Given this size, MinION is well-suited for in field applications and mobile labs. Proof-of-concept studies showed how MinION can be used to detect pathogens with rapid turnaround time, e.g. Ebola or Zika (Faria et al., 2016; Hoenen et al., 2016), enabling real-time diagnostics and public health interventions, such as food safety monitoring or water contamination testing. However, while a number of commercial and open-source software are available for ONT data analysis (de Lannoy et al., 2017), all data processing happens on the cloud, high-performance computing cluster or powerful desktops/laptops, where the analysis is performed. Hence, there is no software built to only use computing resources (RAM, external disk, CPU) available on portable devices, e.g. smartphones and tablets. These devices have different chipset/memory architectures than personal computers or servers, and different operating systems, but in principle existing software could be recompiled to run on such devices. This lack of ONT analytics software that works on a mobile device independently from a wireless connection is a shortcoming to enable true portability. To make an example, MinION could be employed to test for water contamination, e.g. cholera, in a disaster area with power and network outages, or in extreme environments, e.g. space missions (Castro-Wallace et al., 2017). In these settings, where wireless connection is not available or apt to send a process several GB of data over the cloud, even a laptop may not be the preferred choice given weight (e.g. a researcher carrying a backpack with all mobile equipment), power restrictions (backup batteries) and ergonomic needs (swift disinfection of surfaces, touchscreens). In addition, significant improvements in data structures and algorithms have enabled reasonably large-sized datasets to be analyzed and assembled with small amounts of memory and external disk, making the analysis theoretically possible on a portable device (Conway and Bromage, 2011; Milicchio and Prosperi, 2017; Muggli et al., 2019; Nagarajan and Pop, 2013; Pandey et al., 2018; Simpson and Durbin, 2012). In this article, we identified a number of key steps in the analysis of sequence data—from base-calling to genome assembly—and the corresponding software methods built specifically for ONT, recompiled them for mobile devices and benchmarked their performance on smartphones. We chose smartphones for testing because they are architecturally identical to tablets, yet much handier and lighter. Our results were somewhat surprising in that although there are methods specifically designed for ONT data, they are not capable of running on a portable device. Only 11 out of the 23 methods studied were able to be successfully recompiled and ported and only 3 of the remaining methods were able to scale to reasonably sized dataset.
2 Materials and methods
Analytic pipelines for high-throughput sequence data typically use some combination of base-calling, error correction, k-mer counting, mapping to a reference genome and de novo genome assembly (Goodwin et al., 2016). We briefly discuss these steps, and note that, we tested tools that fall into each of these four categories. Base-calling uses the signal produced by the sequencer to determine the nucleotide sequence corresponding to a genome fragment. Next, all unique k-length subsequences (k-mers) and their quantity are identified for a set of sequence reads. These k-mers are not useful per se, but are crucial for many other analytic steps, from read mapping and species annotation to genome assembly. For instance, de novo assemblers based on the de Bruijn graph construct a directed edge for each unique k-mer and label the nodes of each edge with the (k−1)-length prefix and suffix of that k-mer; nodes that have the same label are merged into a single node. In a de Bruijn graph, paths correspond to longer contiguous regions (contigs) of the genome. Another approach to de novo assembly is the overlap-layout-consensus, where an overlap or alignment between reads is computed and a graph representation of this is constructed. Both de Bruijn graph and overlap-layout-consensus approaches are used in modern assemblers due to different advantages they offer (Li et al., 2012). It is worth noting that for both approaches, efficient compression methods that save memory time have been devised (Simpson and Durbin, 2012). Lastly, we note that, there are a number of other types of analysis other than the ones that we tested in this article, including single-nucleotide polymorphism characterization (usually done after reference mapping), functional analysis, structural variation detection or transcriptomics analysis (yet it uses many steps of the reference mapping). For this work, we have chosen the Android operating system and among the ONT software, we selected: Nanocall (David et al., 2017), BasecRAWller (Stoiber and Brown, 2017), Flappie (Runnie) (Technologies, 2019) and Chiron (Teng et al., 2018) for base-calling; LoRMA (Salmela et al., 2017), MarginPolish (ONT, 2019) and Racon (Vaser et al., 2017) for error correction or polishing; DSK (Rizk et al., 2013) for k-mer enumeration; Bowtie2 (Langmead and Slazberg, 2012), BWA (Li and Durbin, 2010), Lambda2 (Hauswedell et al., 2014), Diamond (Buchfink et al., 2015), BLAST (Altschul et al., 1997), Kraken (Wood and Salzberg, 2014) and Minimap2 (Li, 2018) for reference mapping; htslib, SAMtools and BCFtools (Li, 2011; Li et al., 2009) for SAM/BAM analysis; ABruijn (Lin et al., 2016), BCALM2 (Chikhi et al., 2016), Canu (Koren et al., 2017), HINGE (Kamath et al., 2017), SMARTdenovo (Ruan, 2019) and TULIP (Jansen et al., 2017) for de novo assembly, graph compaction and scaffolding. The software choice has been guided primarily by published reviews (de Lannoy et al., 2017). The selection was meant to be comprehensive but also by speed-memory tradeoff, availability of source code and of language compilers for Android. For instance, we did not select the SPAdes genome assembler because of it is very demanding in terms of hardware resources and requires short sequence reads as input—alone or in combination with third-generation sequence reads. Recompiling software for the Android platform, even though a complete toolchain is provided by Google, is often not straightforward. In fact, two major issues are present: the tools of choice must be cross-compilable, and the Android NDK does not always provide all C/C++ standard headers and functions.
2.1 Base-calling
Nanocall is an open-source base-calling software that allows off-line and private analysis of MinION data. It takes as input a set of segmented event sequences stored in ONT-specific FAST5 files and produces nucleotide sequences in FASTQ format. It processes each file separately, performing several rounds of training to obtain valid scale parameters via an expectation–maximization algorithm, and then performs standard Viterbi decoding. Even though it has not been trained specifically for the newer R9.4 MinION’s chemistry it can, in theory, be re-trained. Chiron and BasecRAWller are both base-callers that exploit deep neural network models; while Chiron is batch and optimizes accuracy, BasecRAWller features on-line base-calling during sequencing. Flappie and Runnie are two open-source ONT’s base-callers, both can be directly used on MinION data.
2.2 Error correction
LoRMA is an error correction approach tailored to long reads and high coverage, composed of two main steps: an alignment-free correction based on an iterative de Bruijn graph construction and a polishing step based on long-distance dependencies inferred through multiple alignments. Racon is a parallelized method that corrects post-assembly contigs that did not undergo a consensus step; it is therefore used in conjunction with assemblers like Canu or Minimap2. Racon maps reads to a layout sequence, performing a sliding window and then populating a sliced overlap graph on to which the consensus steps are calculated. MarginPolish is a graph-based assembly refinement tool similar to Racon, takes as input a set of reads and the derived assembly producing a refined assembly by attempting to correct the errors.
2.3 K-mer counting
Counting k-mers is at the base of many high-throughput analytic methods, including de Bruijn graph assembly. Despite the apparent simplicity of this task, sophisticated algorithms and an accurate allocation of resources are required to handle large datasets. DSK uses an out-of-core approach to ensure an efficient use of memory. The algorithm is divided into partitioning and counting. Partitioning is performed in three possible modalities: hashing, frequency-based minimizer and lexicographic-based minimizer. The frequency-based technique has been indicated by the authors as more accurate and more suitable for production of partitions of equal size, and therefore, has been adopted in our tests. The counting is accomplished through hash maps initialized with a constant size of memory. The DSK output consists of a file containing solid k-mers, namely a set of k-mers whose abundance exceeds a given threshold.
2.4 Read alignment
Bowtie2, Minimap2, Lambda, Diamond, Kraken, Blast and BWA are programs that align reads to a reference genome. Bowtie2 uses a full-text index in minute space (FM-index) (Ferragina and Manzini, 2000) of the genome reference sequence based on the Burrows–Wheeler transform (Burrows and Wheeler, 1994) and performs gapped alignment through two stages: first, an ungapped alignment is performed using ‘seed’ substrings from a read (and its reverse complement) using the FM-index, then a gapped extension stage that uses dynamic programing and parallel processing (Langmead et al., 2019) follows. BWA also uses the Burrows–Wheeler transform: it has been designed originally for short sequences, but then extended to handle longer reads—i.e. BWA-MEM. The algorithm is robust to sequencing errors, automatically chooses between local and end-to-end alignments, supports paired-end reads and performs chimeric alignment. Minimap2 has been developed as the successor of BWA-MEM. Minimap2 uses a seed-chain-align procedure based on detection and indexing of minimizers that makes possible the execution of efficient queries to the reference genome obtaining exact matches, i.e. anchors, to the reference. Like Bowtie2 and BWA, Minimap2 also perform a base-level alignment through dynamic programing. Notably, a recent release Minimap2 (Gamaarachchi et al., 2019) introduced an out-of-core option. By storing a split index on disk, a query file is read multiple times and mapped against a batch of target sequences, thus limiting the number of target bases stored in the RAM. This feature leads to a significantly reduced memory consumption, even though more CPU time is required; the memory efficiency also varies when mapping on to a reference genome or on an all-versus-all mapping. Lambda uses a Radix-tree of the queries’ seeds to search in a suffix array built over the subject sequences in order to identify seeds. Then dynamic programing is used during the extension phase. Diamond uses a double-indexing algorithm based on sort-merge join to optimize memory consumption and cache hits. After the indexing part, the two sorted sets are iterated together to identify seeds and then dynamic programing is performed. Kraken is used to assign taxonomic labels to metagenomic DNA sequences. It uses exact k-mer matches and a taxonomy tree to identify matches. Since its approach does not require dynamic programing it is faster than the other alignment methods. After the alignment step, different tools can be used to analyze the output. SAMtools is a set of tools that can be used to interact with SAM, BAM or CRAM files. BCFtools is a suite focused on variance calling. Both belong to the same library HTSlib.
2.5 Genome assembly
BCALM2 is a tool for constructing a compacted de Bruijn graph, which is then used in de novo assembly. The algorithm is divided into three steps: bucketing of k-mers (through the DSK program), compaction of each bucket in parallel mode and, in the last step, k-mers are glued from different buckets with duplicates removed. BCALM2 allows for good balance of memory usage throughout its execution: for instance, for human sequencing data, BCALM2 compacts the de Bruijn graph in roughly an hour using 3 GB of memory. Canu is a revision of Celera, which is a genome assembler specifically developed for long reads. It uses an overlap-layout-consensus strategy with a novel adaptive overlapping strategy (tf-idf weighted MinHash) and a sparse graph build-up that avoids repeat collapsing, with many advantages in terms of coverage requirements and runtime. It also features scaffolding for finishing the genome assembly. ABruijn is another assembler based on the de Bruijn graph that is tailored to long reads. It retains high-frequency k-mers and builds consensus reads, creating a so-called ‘solid’ spectrum, and operates a series of informed choices on the k-mer graph to look for paths supported by k-mer and read solidity. HINGE is an assembler that optimizes repeat resolution by combining the error-resilient overlap-layout-consensus approach with repeat-resolution capabilities of de Bruijn graph assemblers; in detail, it separates repeat regions that are entirely spanned by a read (more easily resolvable) from those that are not, resolving repeats by adding ‘hinges’ emulating a de Bruijn graph repeat collapse. SMARTdenovo performs all-versus-all raw read alignments without error correction and outputs consensus sequences, and recommends its combination with polishing tools like Racon. TULIP tackles the computational burden of all-versus-all read alignments by dividing short ‘seed’ reads from long reads used to connect seed regions. TULIP is fast and memory efficient, although it does not perform any error correction and performance can vary sensibly on the choice of seeds.
2.6 Testing layout
We chose and Android smartphone as testing hardware for a number of reasons, including:
Android smartphones use ARM CPUs, which is the same architecture across all the portable devices like tablets or battery operated programable boards, including edge computing devices.
Android smartphones can be cheaper than the other commercial options and can be easily purchased by every research group.
Offer more functionalities than FPGA devices.
The Android operative system is open source and can be freely customized to fit in bioinformatics pipelines.
We selected MinION sequencing experiment datasets publicly available on NCBI Genbank sequence read archive (SRA), with file sizes ranging from a few hundred MB to 16 GB. We included both single organisms (with a corresponding reference genome, if available) as well as metagenomics samples to explore different genome lengths, base coverage and k-mer spectra. Table 1 shows the sequencing experiments selected for the tests. We calculated with DSK the number of total k-mers (k = 31) and the proportion of unique k-mers: the first one is useful because the file size is not necessarily indicative of genome length or genomic diversity, whilst the second is a rough indicator of errors. The total number of k-mers was highly correlated with the file size (R2=0.99), and on average the percentage of unique k-mers was 71%.
Table 1.
MinION sequencing experiments downloaded from Genbank’s SRA in FASTQ format and used for software benchmarking
| Genbanks SRA Id. | Organism | File size (GB) | Unique k-mers (%) |
|---|---|---|---|
| ERR2900440 | Fermentation M | 0.02 | 53 |
| ERR2900442 | Fermentation M | 0.04 | 49 |
| ERR2900428 | Fermentation M | 0.25 | 35 |
| DRR164915 | Leptotrichia trevisanii | 0.28 | 83 |
| SRR7765365 | Acanthamoeba castellanii | 0.68 | 99 |
| ERR2625614 | Marine V | 0.72 | 57 |
| SRR6037114 | Food M | 1.85 | 42 |
| SRR6037129 | Food M | 2.64 | 31 |
| ERR2564376 | Brassica oleracea | 4.01 | 91 |
| ERR2571299 | Musa schizocarpa | 6.11 | 91 |
| ERR2662964 | Sludge M | 13.23 | 94 |
| SRR5889392 | Oryza coarctata | 13.51 | 97 |
| SRR7762336 | Phaeodactylum tricornutum | 15.10 | 85 |
| ERR2612749 | Magnaporthe oryzae | 16.37 | 81 |
Note: Species with name in italic have a reference genome available.
Each of the aforementioned ONT software, except for Diamond and Kraken that we will discuss later, was run on these datasets on a standard desktop computer as well as on an Android smartphone, if recompilation was successful. For each run, we recorded exit status, CPU time, average/maximum RAM usage and average/maximum CPU temperature. We kept track of the temperature because it can be interpreted as an indicator of the stress to which the device is subjected, also affecting battery consumption and performance. A different test has been designed for Diamond and Kraken. Since Diamond have been specifically designed to be able to analyze database of big size using a customizable quantity of memory we decided to map the metagenomic files of the dataset on UniProt50 (UniProt Consortium, 2018) in order to simulate a real-case scenario. An analogous test has been designed for Kraken but the files have been mapped against the Kraken’s default database built to be of 400 MB.
3 Results
3.1 Software recompilation
In this section, we present the compilation steps, the description of relevant dependencies and the adjustments operated to recompile (successfully or not) the ONT software selected in this work. We used the Android NDK version r17 (Google, 2019a). Nanocall requires zlib (Gailly and Adler, 2019) and HDF5 (HDFgroup, 2019). We gathered platform specific information required by HDF5 from an armv8/arm version of the library and then adjusted for Android. While Flappie has the same external dependencies as Nanocall, it has not been ported because it uses double precision SIMD instructions that are not implemented in Neon. Both Chiron and BasecRAWller require a Python interpreter, Tensorflow (Google, 2019b) and the CUDA platform (Nvidia, 2019). There are Python interpreters for Android and it is possible to build Tensorflow, but CUDA is not available for ARM. Of note, Tensorflow can be run without CUDA but the performance degrades significantly; we decided not to go through this option. We could not recompile LoRMA because it required headers not offered by the NDK Racon (and the SPOA library on which this tool rely) were compiled by including the ‘sse2neon.h’ header in order to convert the Intel AVX instructions to Neon instructions. Note that, this cannot be done for all the tools since the Intel AVX instruction set is wider then Neon. Both DSK and BCALM2 are based on a comprehensive library named GATB (Drezen et al., 2014). This library, written in C++, provides two options for disk storage: standard files and HDF5. Even though we recompiled HDF5 for Nanocall, we decided to remove this dependency from GATB and to adopt the standard file mode in order to simplify maintenance. Bowtie2 has been succesfully compiled since from the last version it uses the SIMDe library to mimic the AVX instruction set when it is not available. BWA could not be recompiled because it required SIMD instructions not available for ARM chipsets. BLAST could not be recompiled because of features incompatibility with the NDK compiler. Minimap2 and Kraken have no external dependencies, other than zlib that is included in the NDK, so recompilation was successful. Canu requires the GNU gcc compiler and no external dependencies. The latest version of the NDK provides only the clang compiler so we were not able to port Canu on Android using the NDK r17. We also tried a previous version of the NDK (r13) that included gcc but the compilation stopped due to a lack of standard headers. One of the ABruijn submodules, GraphMap, requires a recent version of gcc, which is not available and prevented the recompilation; of note, GraphMap also requires a lot of RAM to generate his index during execution. SMARTdenovo requires Streaming SIMD Extensions (SSE), making it not compilable under ARM. For HINGE, we were able to compile the core C/C++ files but then we could not continue because of the CUDA requirement under Tensorflow. TULIP is written in Perl; although there are available Perl interpreters for Android, some of TULIP’s preprocessing steps call BWA. We also ported SAMtools, BCFtools and the underlying library htslib. MarginPolish could not be ported because of the lack of some instructions in the pthread header.
In conclusion, we recompiled successfully 11/23 (47%) of the ONT analytics tools. Table 2 summarizes the tools tested and gives information about dependencies and recompilation issues.
Table 2.
Summary of ONT analytics tools tested, with information about language, dependencies, instruction sets, compilers and recompilation status (with description of issues in case of failure)
| Tool | Language | Link-time dependencies | Instruction sets | Compilers | Recompilation (issue) |
|---|---|---|---|---|---|
| Nanocall | C++ | HDF5; zlib | — | gcc/clang | Successful |
| Flappie | C++ | openblas; hdf5; math | SSE | gcc/clang | Not successful (instruction set) |
| Chiron | Python | Tensorflow | CUDA | — | Not successful (dependencies) |
| BasecRAWller | Python | Tensorflow | CUDA | — | Not successful (dependencies) |
| LoRMA | C/C++ | GATB; zlib; boost | — | gcc/clang | Not successful (headers) |
| Racon | C++ | bioparser; spoa; edlib | Neon | gcc/clang | Successful |
| MarginPolish | C++ | openmp; hts | — | gcc/clang | Not successful (NDK’s implementation of pthread) |
| DSK | C++ | HDF5 | — | gcc/clang | Successful |
| BCALM2 | C++ | HDF5 | — | gcc/clang | Successful |
| Bowtie2 | C++/Python | zlib | — | gcc/clang | Successful |
| BWA | C | zlib | AVX | gcc/clang | Not successful (instruction set) |
| Minimap2 | C | zlib | Neon | gcc/clang | Successful |
| Diamond | C | zlib | — | gcc/clang | Successful |
| Lambda2 | C++ | zlib; SeqAn | — | gcc/clang | Successful |
| Kraken | C++ | — | — | gcc/clang | Successful |
| Canu | C/C++ | openmp; math | — | gcc | Not successful (compiler) |
| Abruijn | C++ | zlib; math; openmp | — | gcc | Not successful (compiler) |
| SMARTdenovo | C | math | SSE | gcc | Not successful (instruction set) |
| HINGE | C/C++/Python | HDF5; boost; Tensorflow | CUDA | gcc/clang | Not successful (dependencies) |
| TULIP | Perl | BWA | — | — | Not successful (dependencies) |
| Samtools | C | zlib | — | gcc/clang | Successful |
| Bcftools | C | zlib | — | gcc/clang | Successful |
| Blast | C++ | — | — | gcc/clang | Not successful (compiler) |
3.2 Software performance
We benchmarked the test layout on a Samsung Galaxy S9+ with 6 GB of memory, an ARM v8 processor Exynos 9810 Octa-core (4 × 2.7 GHz & 4 × 1.7 GHz), 4 GB LPDDR4X RAM and 64GB of storage. We also evaluated performances on a Linux desktop with 32 GB of memory, an Intel® Core i7-6700 CPU @ 3.4 GHz×8.64 GB of internal storage and 1 TB of external storage.
3.2.1. Base-calling
We tested Nanocall on the standard FAST5 files provided with the program. Since FAST5 raw files are split and do not exceed 50 MB in size, Nanocall did not present any issue running on the Android phone.
3.2.2. Assembly and consensus
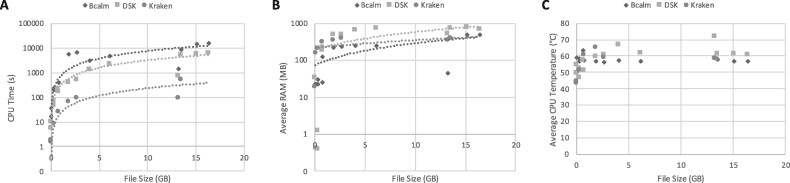
We run DSK and BCALM tools on all files using the desktop and the Android phone. On the phone, DSK processed on average 1 GB in 289 s, whilst BCALM2 took 933 s per GB. The k-mer counting time increased linearly with the file size (R2=0.996), as shown in Figure 1A, and the de Bruijn graph assembly exhibited a similar behavior, although with higher variance (R2=0.783). In terms of RAM usage, both DSK and BCALM2 respected the set RAM limits (800 MB) on average, although a few peaks above the limit (i.e. over 1 GB) were registered. Racon in principle could be run but on our dataset mapping BCALM’s unitigs on the reads file generated SAM files too big to be analyzed.
Fig. 1.
Performance of DSK and BCALM2 on the Android smartphone, all benchmark files (0.02–16 GB), measured by means of CPU time (A), average RAM usage (B) and average temperature (C)
3.2.3. Reference mapping
Minimap2 completed the reference mapping of all the files. The file indexing CPU-times were in the range of 5 s across the whole file size spectrum, while the mapping times increased with the file size (1825 s/GB on average). The average RAM usage for Minimap2 was relatively stable for all file sizes in indexing and increased while mapping, with an average of 23 and 1372 MB, respectively. We also tested Minimap2 all-versus-all mapping on the files without a reference genome but it could not process files larger than 250 MB due to its high RAM consumption, and failed to complete in one of the smaller files. Bowtie2 and Lambda were not able to process files bigger than 250 MB. Both samtools and BCFtools include a variety of different possible analysis so the usability would depend on which analysis and on which input. We tested the command ‘view’ and it worked.
3.2.4. Metagenomic analysis
Kraken was able to complete all the analysis of all the files computing on average 38 MB/s of the input file. Diamond was able to complete the analysis computing on average 1 MB of the input file every minute.
Figure 1 summarizes the performances on mobile showing CPU time (Panel A), average memory consumption (Panel B) and average CPU temperature (Panel C) of DSK, BCALM and Kraken. These tools showed a linear trend on CPU time. On average, BCALM2 and DSK programs reached a CPU temperature of 48° and 49°, with peaks of 69° and 68°. As a comparison, the average operating temperature for a Galaxy S9 is 30°, and the operating system usually shuts off the device when overheating by, although we could not find documentation on the maximum temperature or for how long high temperature can be sustained.
3.2.5. Desktop
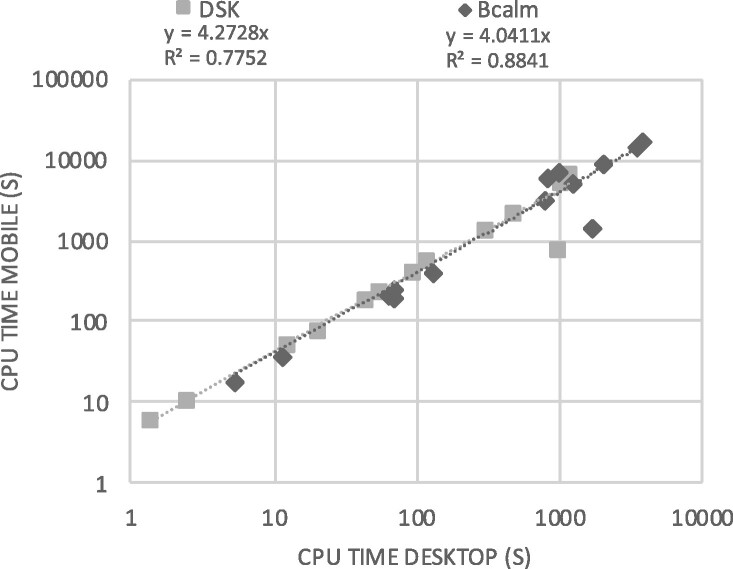
In Figure 2, we plotted the CPU time on the phone versus the CPU time on the desktop by DSK and BCALM2 to see if processing times on the different architectures were correlated. For DSK, they were linearly correlated (R2=0.7752) with the desktop being on average four times faster; BCALM2 exhibited also good linear correlation (R2=0.8841) and it was four times faster on the desktop, although with higher variation.
Fig. 2.
Comparison of data processing times by DSK and BCALM2 on the Android smartphone versus the desktop execution for all benchmark files (0.004–16GB). Note that, for smartphone runs, an 800-MB ram limit have been imposed on both DSK and BCALM
4 Discussion
In this work, we analyzed the portability of ONT analytics on to mobile architectures (an Android smartphone in our case) by selecting, recompiling and benchmarking a set of commonly used software, from base-calling to genome assembly. We were able to compile only 11 out of 23 tools. The reason behind the difficulties encountered in porting the software includes lack of standard headers, compilers and unsupported library dependencies or instruction sets. Maintenance is a big problem in porting, because many adaptations are necessary during the compilation and they challenge reproducibility from version to version. Unfortunately Canu, which is in the ONT assembly pipeline endorsed by Oxford’s Nanopore (Jain et al., 2018), could not be compiled and tested on the smartphone. The programs written in Python (Chiron, BasecRAWller and HINGE) could have been in principle ported, but the Tensorflow library would have not made use of the CUDA acceleration. RAM usage made Minimap2 all-versus-all unusable even at relatively small file sizes (250 MB and up), which led only Minimap2 reference mapping, Kraken, DSK, BCALM2 to run successfully on all files up to 16 GB. Moreover, Diamond was able to map small files (i.e. 400 MB) to the entire UniProt50 database. It has to be noticed that some of this softwares partition the input in order to obey a memory constraint, usually through an informed guess, but this could lead to unusual RAM usage peaks that may pose problems if they get close to the maximum usable RAM in absence of an efficient out-of-core handling. In terms of CPU time, DSK, BCALM2, Kraken and Minimap2 mapping scaled linearly with the desktop time, which is a reassuring result, and the overall timing of data processing is bearable if thought for real-time settings. MinION Flow Cells output data from a few GB to 20 GB, and in this work 1 GB was processed in 5–10 min by DSK/BCALM2, and in less than a minute by Kraken, on the phone. It is expected that newer chemistry for MinION will allow for 40 GB per flow cell. Since a MinION sequencing experiment can take ∼40 min including sample preparation, faster methods to allow portable analytics in real time will be needed. A more serious concern is that CPU temperature rose dramatically for all programs: apart from system-forced shutdowns that would prevent completion of analyses, high temperature would also affect battery, device and CPU life. It would be difficult to handle the device and it could lead to a fire hazard. These issues are non-negligible in mobile lab settings. This work has limitations: first, we tested only a relatively small number of software, which may not be fully representative of the entire ONT software available on the market; second, we did not altered the source code other than providing instruction set conversion headers; third, we used only the Android operating system, while porting could be different and lead to different results using Apple’s iOS; finally, we did not analyze battery consumption. In conclusion, the software scenario for ONT analytics does not seem to be suitable to enable true portability now: we are not there yet. Development of new tools tailored to ONT should be carried on with special attention to mobile architectures: if not purposely built for ARM-based system-on-chip, at least the code writing should be attentive to portable headers, library dependencies and usage of instruction sets that are also available for Android or iOS. Further development should also focus on optimization in resource-constrained settings, which include not only RAM, but also CPU-temperature throttling. There are preliminary approaches to develop ONT analytic tools specifically tailored to mobile architectures but they are at a very nascent stage, e.g. NanoPAL, a generic programing library that exploits out-of-core approaches (Milicchio et al., 2016, 2018). Besides command-line development, another critical area for ONT portable analytics is expected to be the development of graphical user interfaces and visualization, given the small screen size of portable devices. Existing end-to-end and touch-app solutions like Illumina’s BaseSpace (Illumina, 2019) or ONT MinION Mk1C might be a starting point but they are not open source or free, and it may be difficult to develop pipelines within their systems, unless the companies decide to do so. As a final message, testing compilation on mobile architectures and provide binaries could be a good recommendation for ONT software developers, especially with the upcoming release of the SmidgION sequencers that directly plug into a smartphone.
Funding
This work was supported by the National Institutes of Health NIAID [1R01AI141810-01 to M.P. and C.B.]; and by the UF Moonshot initiative.
Conflict of Interest: none declared.
Contributor Information
Marco Oliva, Department of Engineering, Roma Tre University, Rome, Italy; Department of Computer and Information Science and Engineering.
Franco Milicchio, Department of Engineering, Roma Tre University, Rome, Italy.
Kaden King, Department of Computer and Information Science and Engineering.
Grace Benson, Department of Computer and Information Science and Engineering.
Christina Boucher, Department of Computer and Information Science and Engineering.
Mattia Prosperi, Department of Epidemiology, University of Florida, Gainesville, FL 32610, USA.
References
- Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B. et al. (2015) Fast and sensitive protein alignment using diamond. Nat. Methods, 12, 59–60. [DOI] [PubMed] [Google Scholar]
- Burrows M., Wheeler D.J. (1994) A block-sorting lossless data compression algorithm. Technical report.
- Castro-Wallace S.L. et al. (2017) Nanopore DNA sequencing and genome assembly on the international space station. Sci. Rep., 7, 18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi R. et al. (2016) Compacting de Bruijn graphs from sequencing data quickly and in low memory. Bioinformatics, 32, i201–i208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T.C., Bromage A.J. (2011) Succinct data structures for assembling large genomes. Bioinformatics, 27, 479–486. [DOI] [PubMed] [Google Scholar]
- David M. et al. (2017) Nanocall: an open source basecaller for Oxford Nanopore sequencing data. Bioinformatics, 33, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lannoy C. et al. (2017) The long reads ahead: de novo genome assembly using the MinION. F1000Res, 6, 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drezen E. et al. (2014) GATB: genome assembly & analysis tool box. Bioinformatics, 30, 2959–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R. et al. (2016) Mobile real-time surveillance of Zika virus in Brazil. Genome Med., 8, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferragina P., Manzini G. (2000) Opportunistic data structures with applications. In: Proceedings of Annual Symposium on Foundations of Computer Science, Redondo Beach, CA. pp. 390–398.
- Gailly J., Adler M. (2019) zlib. https://www.zlib.net (1 April 2020, date last accessed).
- Gamaarachchi H. et al. (2019) Featherweight long read alignment using partitioned reference indexes. Sci. Rep., 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S. et al. (2016) Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet., 17, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Google (2019. a) Native Development Kit. https://developer.android.com/ndk (1 April 2020, date last accessed).
- Google (2019. b) Tensorflow. https://www.tensorflow.org (1 April 2020, date last accessed).
- Hauswedell H. et al. (2014) Lambda: the local aligner for massive biological data. Bioinformatics, 30, i349–i355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HDFgroup (2019) hdf5. https://www.hdfgroup.org (1 April 2020, date last accessed).
- Hoenen T. et al. (2016) Nanopore sequencing as a rapidly deployable Ebola outbreak tool. Emerg. Infect. Dis., 22, 331–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illumina (2019) Basesspace. https://basespace.illumina.com (1 April 2020, date last accessed).
- Jain M. et al. (2016) The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol., 17, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M. et al. (2018) Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol., 36, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen H.J. et al. (2017) Rapid de novo assembly of the European eel genome from nanopore sequencing reads. Sci. Rep., 7, 7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath G.M. et al. (2017) HINGE: long-read assembly achieves optimal repeat resolution. Genome Res., 27, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S. et al. (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res., 27, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. et al. (2019) Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics, 35, 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics, 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics, 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. ; 1000 Genome Project Data Processing Subgroup. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. et al. (2012) Comparison of the two major classes of assembly algorithms: overlap-layout-consensus and de-Bruijn-graph. Brief. Funct. Genomics, 11, 25–37. [DOI] [PubMed] [Google Scholar]
- Lin Y. et al. (2016) Assembly of long error-prone reads using de Bruijn graphs. Proc. Natl. Acad. Sci. USA, 113, E8396–E8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low L., Tammi M.T. (2017) Introduction to Next Generation Sequencing Technologies. World Scientific, Singapore. [Google Scholar]
- Ma X. et al. (2017) Evaluation of oxford nanopore MinIONTM sequencing for 16S rRNA microbiome characterization. BioRxiv, 099960. [Google Scholar]
- Milicchio F., Prosperi M. (2017) Efficient data structures for mobile de novo genome assembly by third-generation sequencing. Procedia Comput. Sci., 110, 440–447. [Google Scholar]
- Milicchio F. et al. (2016) High-performance data structures for de novo assembly of genomes: cache oblivious generic programming. In: Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Seattle, WA. pp. 657–662.
- Milicchio F. et al. (2018) Third-generation sequencing data analytics on mobile devices: cache oblivious and out-of-core approaches as a proof-of-concept. Procedia Comput. Sci., 134, 219–226. [Google Scholar]
- Muggli M.D. et al. (2019) Building large updatable colored de Bruijn graphs via merging. Bioinformatics 35, i51–i60. [DOI] [PMC free article] [PubMed]
- Nagarajan N., Pop M. (2013) Sequence assembly demystified. Nat Rev Genet., 14, 157–67. [DOI] [PubMed]
- Nvidia (2019) Cuda. https://developer.nvidia.com (1 April 2020, date last accessed).
- ONT (2019) Marginpolish. https://github.com/UCSC-nanopore-cgl/MarginPolish (1 April 2020, date last accessed).
- Pandey P. et al. (2018) Mantis: a fast, small, and exact large-scale sequence-search index. Cell Syst., 7, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk G. et al. (2013) DSK: k-mer counting with very low memory usage. Bioinformatics, 29, 652–653. [DOI] [PubMed] [Google Scholar]
- Ruan J. (2019) Smartdenovo. https://github.com/ruanjue/smartdenovo (1 April 2020, date last accessed).
- Salmela L. et al. (2017) Accurate self-correction of errors in long reads using de Bruijn graphs. Bioinformatics, 33, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.T., Durbin R. (2012) Efficient de novo assembly of large genomes using compressed data structures. Genome Res., 22, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoiber M., Brown J. (2017) Basecrawller: streaming nanopore basecalling directly from raw signal. bioRxiv. 133058. doi: 10.1101/133058. [Google Scholar]
- Technologies O.N. (2019) Flappie. https://github.com/nanoporetech/flappie (1 April 2020, date last accessed).
- Teng H. et al. (2018) Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning. Gigascience, 7. doi: 10.1093/gigascience/giy037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2018) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res., 47, D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R. et al. (2017) Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res., 27, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol., 15, R46. [DOI] [PMC free article] [PubMed] [Google Scholar]