Figure 1.
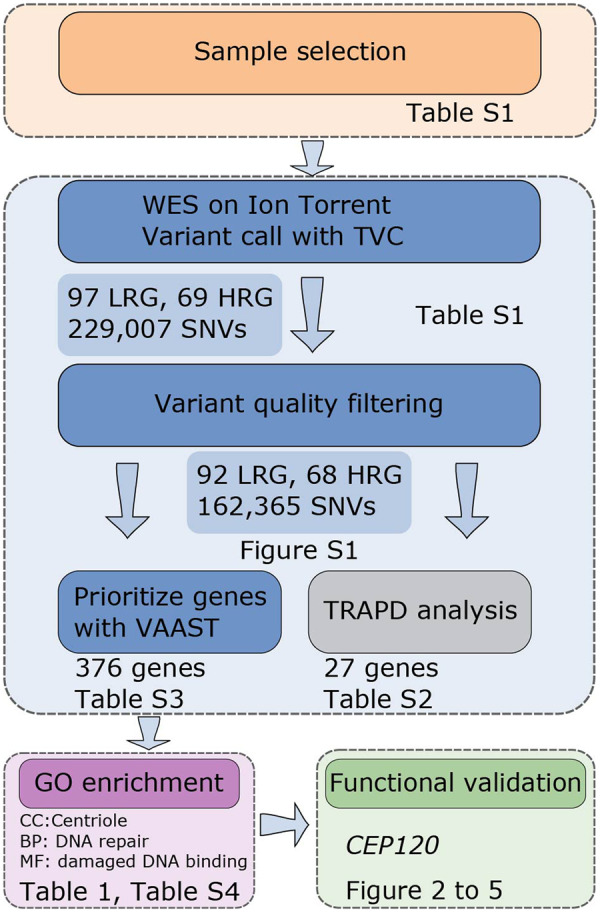
Overall strategy to identify candidate aneuploidy genes in human. Selected patients were subjected to exome sequencing using the Ion Torrent platform. Variants were identified using Torrent Variant Caller (TVC) software and high-quality single-nucleotide variant (SNV) were used for association analysis. Mutation burden analysis was performed with TRAPD (Testing Rare vAriants using Public Data) and VAAST (Variant Annotation, Analysis and Search Tool). Genes identified by VAAST were used for gene ontology (GO) enrichment and pathway analysis. Centrosomal protein 120 (CEP120) was selected for functional studies in a mouse oocyte model.
