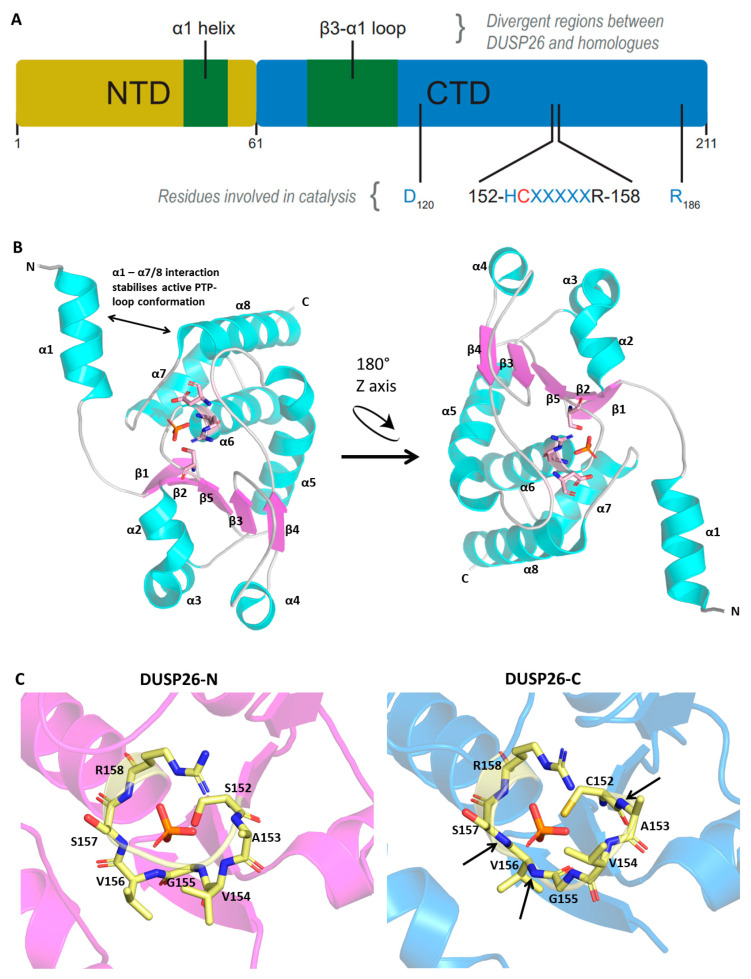
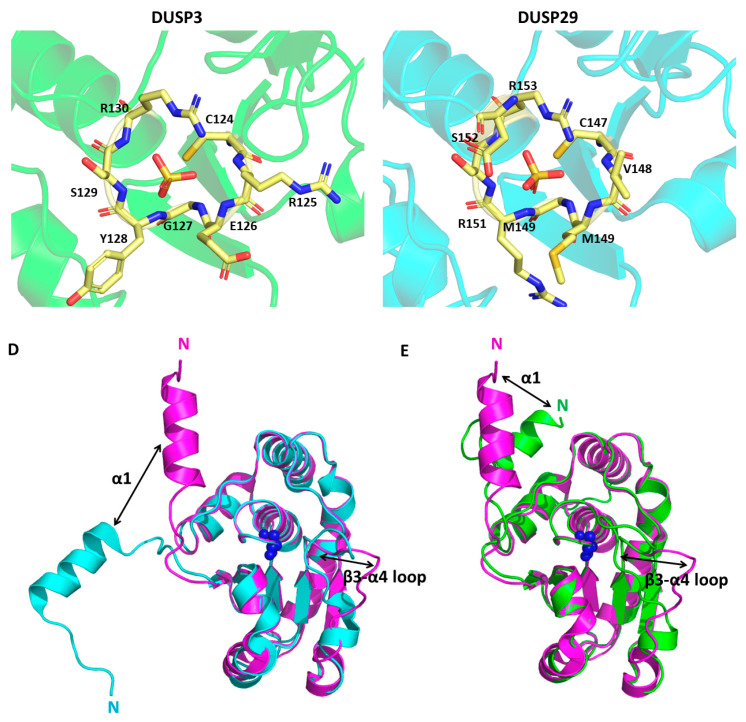
Figure 2.
DUSP26 structure and comparison to homologous proteins. Protein Data Bank (PDB) file retrieved from the RCSB PDB Database and loaded into the Pymol software. (A) Schematic of protein domain structure depicting N- and C-terminal domains. (B) Crystal structure of DUSP26-N monomer presented with the catalytic triad surrounding the phosphate ion. (C) PTP-loop residues of DUSP26-N, DUSP26-C, DUSP3 and DUSP29 surrounding a phosphate (DUSP26-N and DUSP26C) or sulphate (DUSP3 and DUSP29) ion. The phosphate ion residing in the DUSP26-C PTP-loop has been superimposed from the DUSP26-N structure. Black arrows identify amide bonds in the DUSP26-C PTP-loop that are not pointing towards the phosphate ion centre. Structural alignment of DUSP26-N (magenta) with (D) DUSP3 (cyan) and (E) DUSP29 (green). Disparate structural regions are depicted by double-edged arrows and the catalytic Cys152 depicted as blue spheres. PDB codes: DUSP26-N, 5GTJ; DUSP26-C, 2E0T; DUSP3, 1VHR; DUSP29, 2Y96. Figures adapted from [43].