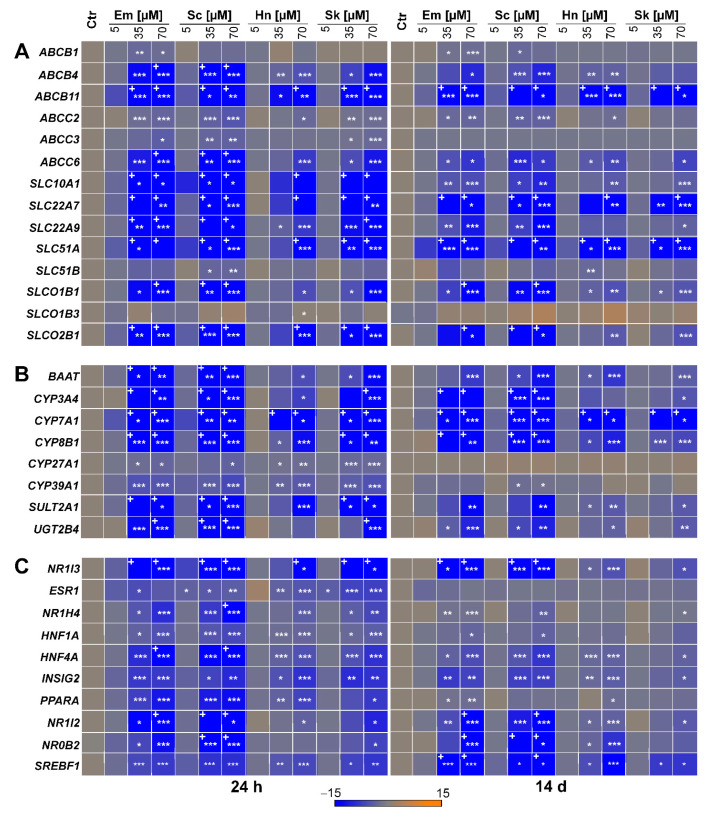
Figure 2.
PA-dependent alterations of bile acid homeostasis-associated gene expressions ((A): transporters, (B): enzymes, (C): nuclear receptors) in HepaRG cells. HepaRG cells were seeded and cultivated as described in the material and methods section. Subsequently, the cells were incubated for 24 h or 14 days with 5, 35, or 70 µM of PA or the solvent (Ctr; 1.7% DMSO and 0.7% ACN). The total RNA was isolated and transcribed into cDNA. Expression analysis was performed by qRT-PCR. Gene expression of the target gene was normalized to ACTB and referred to solvent control to obtain relative expression (2−ΔΔCt method). The heat map shows the relative expression values for up- and downregulation as positive and negative fold changes (means of three independent experiments with three replicates each). For better comparability, only the fold change range between −15 and 15 is shown. Values exceeding this range are marked with +. Statistical differences were evaluated using one-way ANOVA followed by Dunnett’s test: * p < 0.05, ** p < 0.005, *** p < 0.001. Em, echimidine; Sc, senecionine; Hn, heliotrine; Sk, senkirkine.