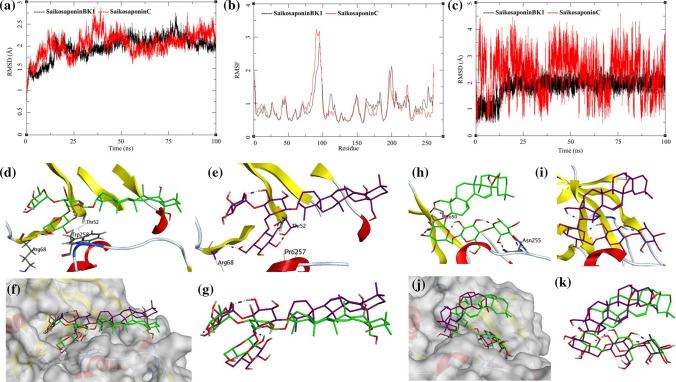
Fig. 7.
Trajectory analysis for NADPH-Oxidase 5 (NOX5) (PDB: 5O0X) bound to Saikosaponin-BK1 and Saikosaponin-C; a root-mean-square deviation (RMSD), b root-mean-square fluctuations per amino acid (aa) (RMSF) and c root-mean-square deviation for each ligand (Lig-RMSD). Interaction analysis of the NADPH-oxidase 5 (NOX5) (PDB: 5O0X) bound to Saikosaponin-BK1 during the molecular dynamics simulation; d equilibrated structure of Saikosaponin-BK1 (green) bound to the NOX5 before MDS production phase; e Saikosaponin-BK1 (purple) bound to the NOX5 post-MDS production phase; f receptor surface analysis; g ligand analysis. Interaction analysis of the NADPH-Oxidase 5 (NOX5) (PDB: 5O0X) bound to Saikosaponin-C during the molecular dynamics simulation; h equilibrated structure of Saikosaponin-C (green) bound to the NOX5 before MDS production phase; i Saikosaponin-C (purple) bound to the NOX5 post-MDS production phase; j receptor surface analysis; k ligand analysis