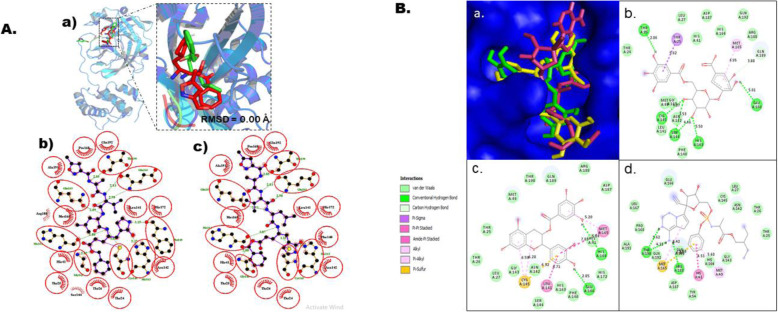
Fig. 2.
Molecular docking studies of gallic acid derivatives against main protease (Mpro) of SARS-Cov-2. a Protocol validation of molecular docking experiment using AutoDock Vina, PyMOL, and LigPlot+. (a) Comparison of binding modes for re-docked ligand (red) vs. co-crystallized ligand (green) shown as stick representation. Amino acid residues interaction with (b) co-crystalized and (c) re-docked ligand accomplished in LigPlot+. b Binding mode and molecular interaction of hit ligands with Mpro. (a) Surface representation of Mpro (PDB: 6 LU7) show the binding mode of docked 3-O-(6-galloylglucoside) (yellow), epicatechin gallate (green), and remdesivir (pink). 2D interaction of (b) 3-O-(6-galloylglucoside), (c) epicatechin gallate, and (d) remdesivir