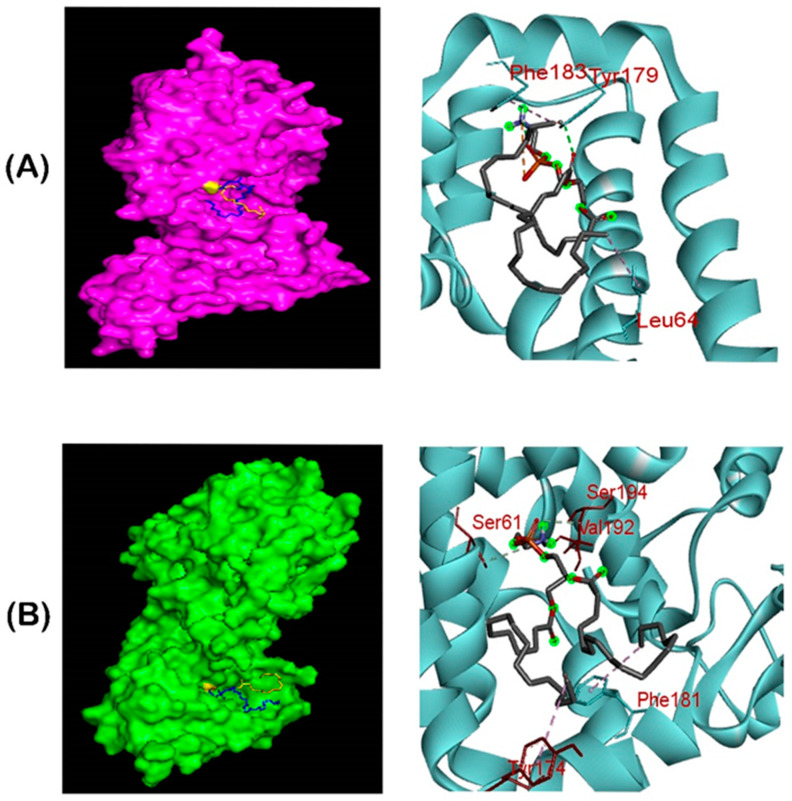
Figure 6.
Ligand–binding interaction pattern of phosphatidylethanolamine substrate with colistin resistance MCR-1 and LptA. The modeled ribbon structure for PE substrate with MCR-1 protein. The ribbon structure was given via PyMol software. In both (A,B) cases, the substrates tend to bind in the groove of MCR-1 and LptA, mostly spanning the 175–195 region, in which Phe, Tyr and Ser residues were abundantly found in the substrate binding region for phosphatidylethanolamine interaction. (B) Ligand–binding interaction revealed that both MCR-1 and LptA proteins exhibited similar localization of phosphatidylethanolamine binding sites (SAUVM MCR-1: Leu 64, Tyr 179, Phe 183; LptA: Ser 61, Tyr 174, Phe 181, val 192, Ser 194).