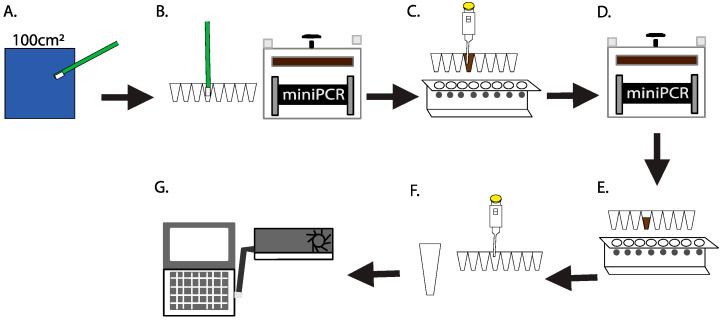
Figure 1.
Workflow of the in-situ culture-independent, swab-to-sequencer process for extreme environment microbial profiling. (A) Swabbing of a 100 cm2 area. (B) Insertion of the swab head directly into lysis buffer in a PCR tube and placement into miniPCR for DNA extraction. (C) Bead-based DNA purification (D) DNA amplification within miniPCR. (E) Bead-based PCR amplicon purification. (F) Addition of sequencing control DNA and ONT-based rapid adaptor library preparation. (G) Flow cell loading and MinION sequencing.