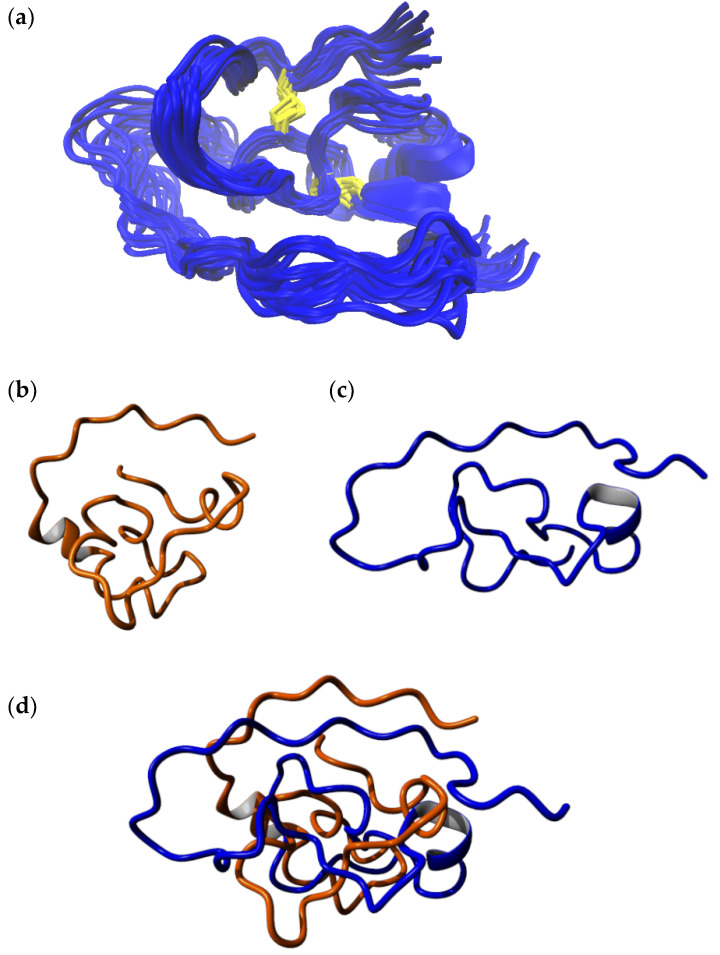
Figure 5.
(a) The final 20 snapshots of the 300 ns MD simulation of the full-length model of isomer B[C19S,C25S] [29]. (b) Computationally derived structure of the same peptide analogue from an earlier study [29]. (c) Final snapshot from the 300 ns simulation from the current study. (d) Structural alignment of the final structure from the 300 ns MD simulation from the current study (blue) with the computationally derived structure of the same peptide (orange) from an earlier study [29]. The alignment indicates that the two models essentially have varying structures while the underlying fold is preserved. The computationally derived structure from earlier work has a more compact fold owing to the fact that it was created based on its three-disulfide-bonded parent, where residues Cys19 and Cys25 were linked by a disulfide bond. A fair alignment between the trailing end of the C-terminal segment is observed. The alignment was carried out using the MUSTANG algorithm [36].