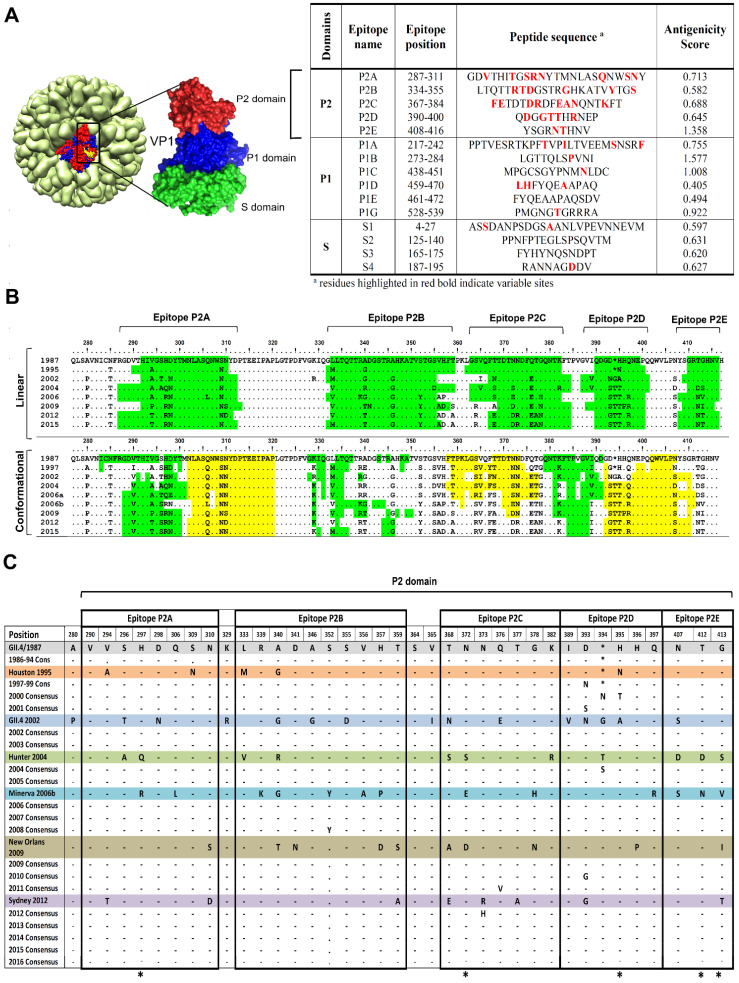
Figure 1.
Predicted B-cell epitopes in NoV GII.4 variants and their evolutionary pattern. (A) Linear B-cell epitopes were predicted within norovirus Sydney GII.4 capsid protein using IEDB, Epitopia27 and BepiPred prediction tools: epitopes with antigenicity index higher that 0.4 were considered as antigen. NoV crystal structure (1IHM) was taken from protein data bank and structure was generated with chimera tool. (B) Evolutionary pattern of the predicted linear and conformation epitopes was determined within the GII4 variants connected with norovirus outbreaks. Yellow and green colors have been used to differentiate closed neighboring conformational epitopes (C) Columns of heterogenecity within P2 domain. Respective variants constituted yearly consensus sequences till they were replaced by the next variant. Positively selected sites are labelled with an asterisk below the columns.