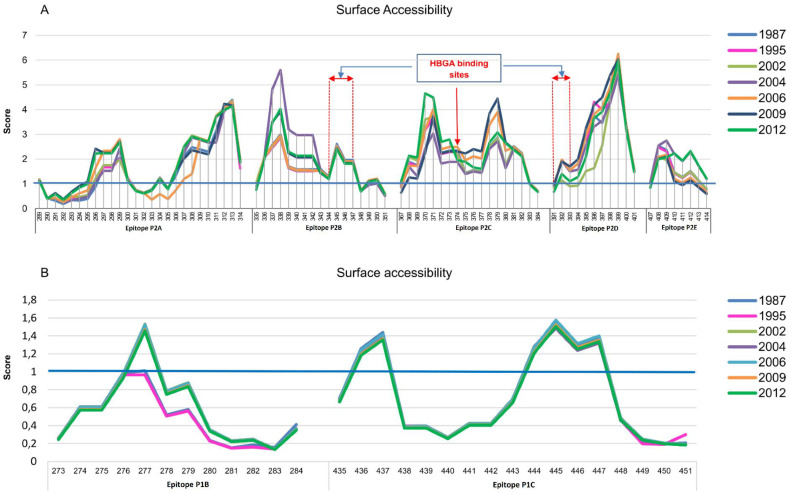
Figure 2.
Surface accessibility of amino acid residues within epitopes sites. Capsid protein from norovirus variants responsible for outbreaks were assessed for changes in surface accessibility within epitopes sites using the Emini Surface Accessibility Prediction tool. (A) Epitopes within the P2 domain showed changes in a mutation-related manner. HBGA binding sites are located within epitopes P2B, P2C and P2D; (B) Epitope sites in the surface exposed sites of P1 domain (P1B and P1C) were stable over years.