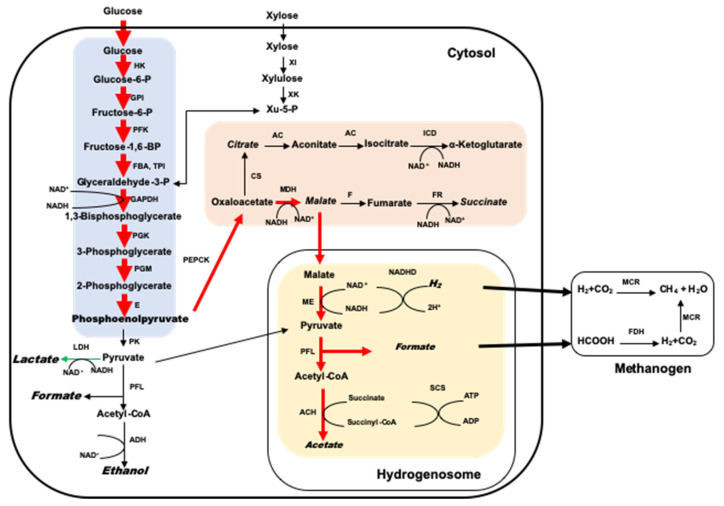
Figure 3.
Proposed metabolic pathway for glucose by anaerobic fungi co-cultured with methanogens. The red and green arrows indicate stimulated and inhibited pathways. The proposed metabolites are indicated in italics. AC, aconitase; ACH, acetyl-CoA hydrolase; ADH, alkoholdehydrogenase; CS, citrate synthase; E, enolase; F, fumarase; FBA, fructosebisphosphate aldolase; FDH, formate dehydrogenase; FR, fumarate reductase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GPI, glucose-6-phosphate isomerase; HK, hexokinase; ICD, isocitrate dehydrogenase; LDH, lactatedehydrogenase; MCR, methyl coenzyme-M reductase; MDH, malate dehydrogenase; ME, malic enzyme; NADHD, NADH dehydrogenase; SCS, succinyl-CoA synthetase; TPI, triosephosphate isomerase; XK, xylulokinase; XI, xylose isomerase; PFK, phosphofructokinase; PFL, pyruvate formate lyase; PGK, 3-phosphoglycerate kinase; PGM, phosphoglucomutase; PK, pyruvate kinase; PEPCK, phosphoenolpyruvate carboxykinase.