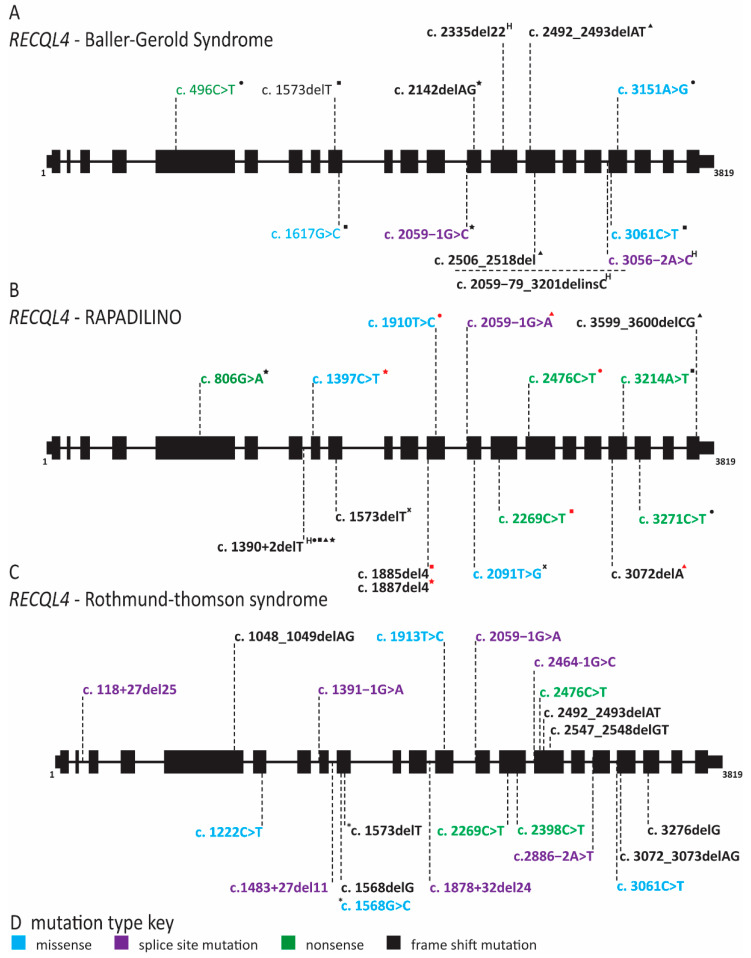
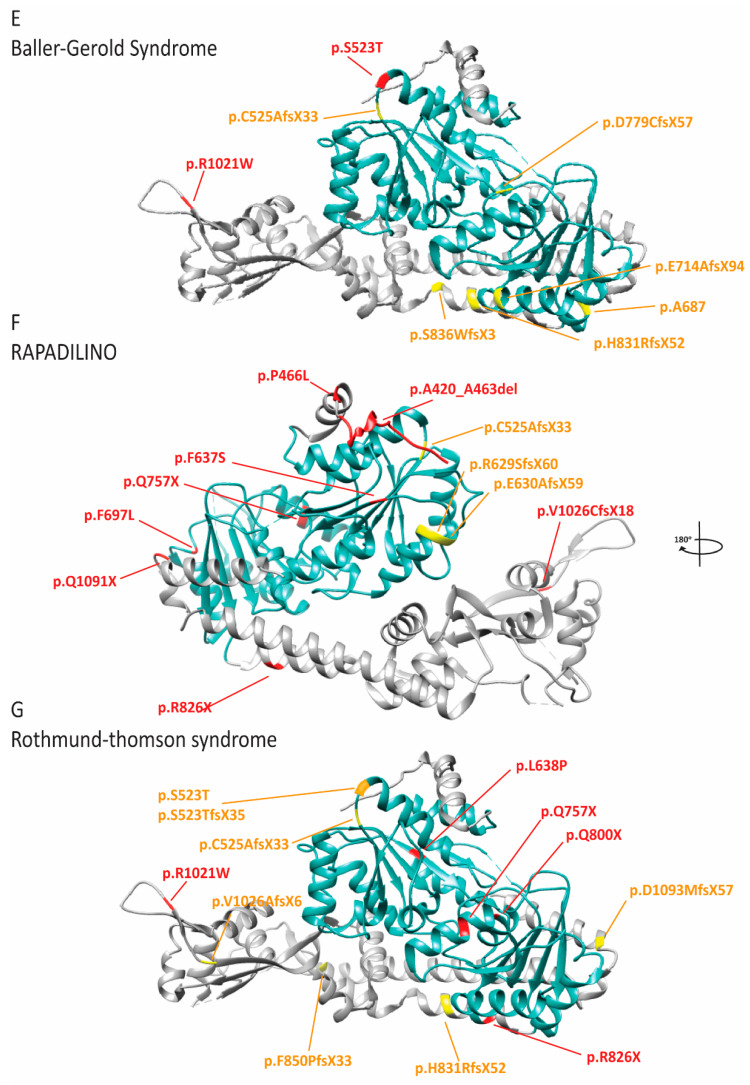
Figure 12.
Pathogenic variants in RECQL4 are associated with three different syndromes, Rothmund–Thomson (RTS), Baller–Gerold (BGS) and RAPADILINO (RAdial hy-po/aplasia, PAellae hypo/aplasia and cleft PAlate, DIarrhea and DIslocated joint, LIttle size and LImb malformation, NOse slender and NOrmal intelligence). Mutations of RECQL4 in BGS, RAPADILINO and RTS are depicted on a schematic of the gene and crystal structure of Homo sapiens RECQL4. For a complete list of mutations and associated protein changes, see Supplementary Table S1. The structure was generated using pdb file 5lst and the Chimera program (http://www.cgl.ucsf.edu/chimera). Exon and intron schematics were generated with the UCSD genome browser. Reference sequence is RECQL4 NM_004260.4. Schematic of RECQL4 gene for (A) BGS and (B) RAPADILINO is depicted with exons as black boxes and introns as horizontal lines. Mutations are mapped to the appropriate gene regions. “H” superscript indicates a homozygous mutation. Compound heterozygous mutation pairs are indicated by superscript symbols. Each mutation combination is depicted, and certain alleles may be present in multiple combinations. The frequency of each allele in the affected population is not indicated. (C) Mutations that occur in multiple individuals with RTS are depicted. “*” indicates that these mutations occur in cis, c.1573delT does occur alone in some individuals (D) For each gene, the color of the mutations indicates the type of mutation according to this key. Crystal structure of human RECQL4 with the helicase domain highlighted in blue, missense and nonsense mutations highlighted in red and frameshift mutations highlighted in orange for (E) BGS, (F) RAPADILINO and (G) RTS. For RTS, one amino acid indicated in orange on the ribbon structure has two mutations associated with it. Mutations that do not have corresponding amino acids in the structure or resulted in abnormal splicing are not depicted.