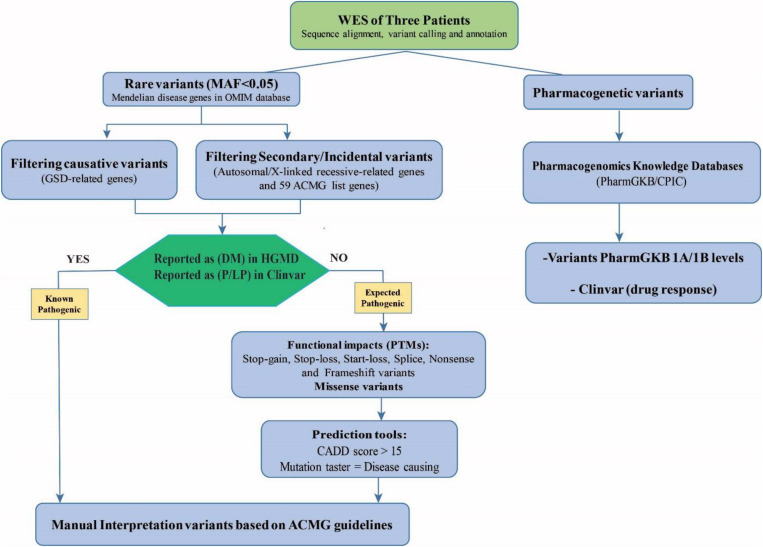
FIGURE 1.
The workflow filtering of the causative variants and the secondary actionable findings in the patients for the WES results. The assessment of WES data consists of filtering for rare known pathogenic (P) and likely pathogenic (LP) variants in genes in the OMIM database including the GSD-associated genes, the ACMG’s list of 59 genes, and other genes associated with mendelian diseases (autosomal/X-linked recessive-related genes). For prioritizing expected pathogenic variants, protein-truncating variants (PTMs) and missense variants were selected and bioinformatics prediction tools were used to evaluate the potential impact and pathogenicity of the candidate variants (CADD score > 15 and mutation taster = disease causing). The identified variants were categorized and interpreted according to the ACMG-AMP 2015 Standards and Guidelines. Besides, secondary pharmacogenetic variants in the Pharmacogenomics Knowledgebase (PharmGKB class IA/IB) and ClinVar database were explored.