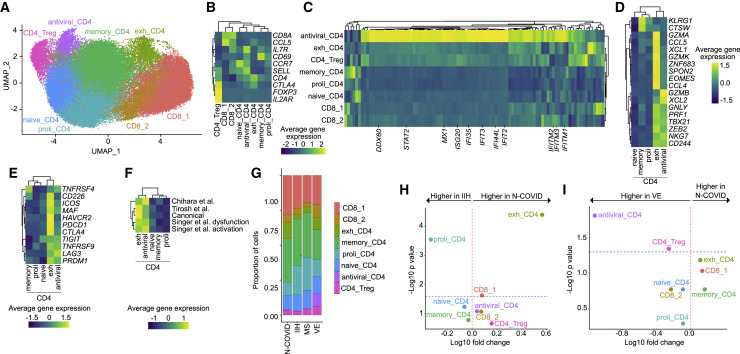
Figure 3.
Exhausted CD4+ T Cells Expand in the CSF in Neuro-COVID
(A) UMAP plot depicting 8 color-coded T cell subclusters from Neuro-COVID (N-COVID; n = 8), 9 idiopathic intracranial hypertension (IIH) (n = 9), multiple sclerosis (MS) (n = 9), and viral encephalitis (VE) (n = 5) patients; in total 61,642 single-cell transcriptomes.
(B) Average gene expression of canonical T cell markers in T cell subclusters.
(C) Average gene expression of interferon-stimulated genes obtained from Wilk et al. (Wilk et al., 2020) in T cell subclusters. The complete list of genes is provided in Table S4.
(D) Average gene expression of cytotoxicity markers in CD4+ clusters obtained from Patil et al. (Patil et al., 2018).
(E) Average gene expression of canonical T cell exhaustion markers in CD4+ clusters.
(F) Average gene expression of T cell exhaustion gene sets from Singer et al. (Singer et al., 2016), Tirosh et al. (Tirosh et al., 2016), Chihara et al. (Chihara et al., 2018), and canonical exhaustion markers in CD4+ T cell clusters. The complete gene sets are provided in Table S4.
(G) Proportions of T cells split by diagnosis.
(H and I) Changes of T cell subcluster abundances in Neuro-COVID (n = 8) versus IIH (n = 9) (H) and Neuro-COVID (n = 8) versus VE (n = 5) (I). Logarithmic fold change of cluster abundance is plotted against negative logarithmic p value (two-sided Wilcoxon’s rank-sum test). The horizontal dashed line represents the significance threshold (p = 0.05).
Abbreviations: proli_CD4, proliferating CD4+ T cells; memory_CD4, memory-like CD4+ T cells; exh_CD4, exhausted CD4+ T cells; CD8, CD8+ T cells; CD4_Treg, regulatory CD4+ T cells.