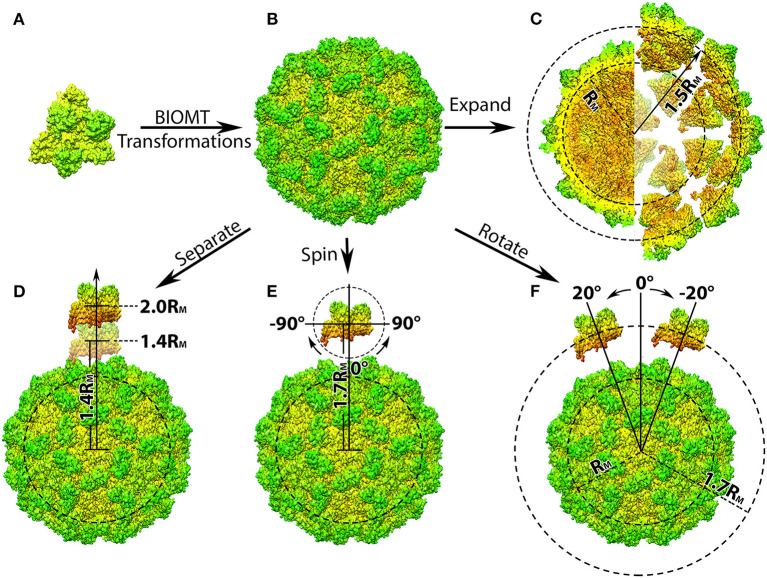
Figure 1.
Structure manipulation of the capsid of Turnip Crinkle Virus (TCV). (A) Individual capsomer of TCV with the three type of protein subunits labeled on the side (PDB ID 3ZX8); (B) the whole TCV capsid generated using the capsid generation tool; Penal (C–F) are demonstration of structure manipulation where: (C) the native TCV capsid is expended by 0.7RM. The central section of the native capsid (left) and the expended capsid (right) are shown as their radius labeled in black, respectively; (D) one capsomer is detached from the rest of the capsid by 0.4 to 1.0RM, as its distance to the mass center of the whole capsid increases to 1.4RM and 2.0RM; (E) one capsomer is first detached by 0.7RM and then spun from −90Å to 90Å in the xy-plane (around z-axis); (F) after detached by 0.7RM, the capsomer is rotated around the capsid from −20Å to 20Å with respect to the mass center of the whole capsid. In all panels, the capsomers and capsids are shown in their density map generated using Chimera (Pettersen et al., 2004; Goddard et al., 2007) and colored by radius from red to green.