Highlights
-
•
A novel grey Richards model GERM(1,1,) is proposed.
-
•
The optimal nonlinear terms and background value of the novel model are determined by Genetic algorithm.
-
•
The comparative study shows that the new model is superior to the other seven benchmark models.
-
•
The predict the daily number of new confirmed cases of COVID-19 of four regions are projected.
Keywords: Grey prediction model; COVID-19; Traditional Richards model; Genetic algorithm optimization; GERM(1,1,)
Abstract
In 2020, a new type of coronavirus is in the global pandemic. Now, the number of infected patients is increasing. The trend of the epidemic has attracted global attention. Based on the traditional Richards model and the differential information principle in grey prediction model, this paper uses the modified grey action quantity to propose a new grey prediction model for infectious diseases. This model weakens the dependence of the Richards model on single-peak and saturated S-shaped data, making Richards model more applicable, and uses genetic algorithm to optimize the nonlinear terms and the background value. To illustrate the effectiveness of the model, groups of slowly growing small-sample and large-sample data are selected for simulation experiments. Results of eight evaluation indexes show that the new model is better than the traditional GM(1,1) and grey Richards model. Finally, this model is applied to China, Italy, Britain and Russia. The results show that the new model is better than the other 7 models. Therefore, this model can effectively predict the number of daily new confirmed cases of COVID-19, and provide important prediction information for the formulation of epidemic prevention policies.
1. Introduction
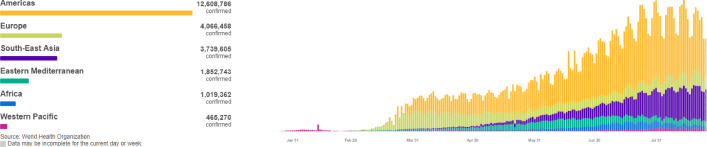
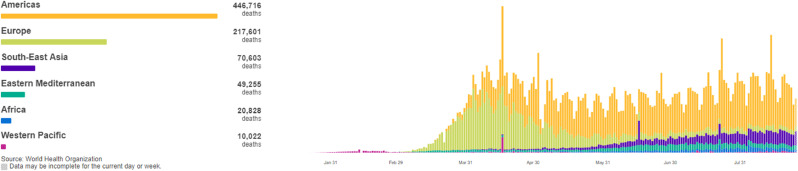
In 2020, a new type of coronavirus (COVID-19) has broken out across the world. The World Health Organization (WHO) has announced that COVID-19 has entered a global pandemic. Till now, data from the WHO [1] shows that more than 20 million confirmed cases of COVID-19 have been reported globally. After breaking the 10 million mark on June 28, the number of confirmed cases worldwide has doubled in about six weeks. In recent months, the change trend of six regions worldwide is shown in Fig. 1 [1]. Fig. 1 shows that the center of the epidemic has moved from the Western Pacific region to Europe at the beginning, and is currently staying in the Americas. The daily number of newly diagnosed patients in the six regions shows a fluctuating S-shaped trend. According to the death toll trend chart in Fig. 2 [1], except for the Americas, the death toll in other regions is basically under control at this stage. During the virus outbreak stage, the death rate in individual countries exceeded 10%, and the death rate in most countries between 5% and 10%, the total death toll at this stage has exceeded 700,000. The global epidemic is still severe, so the worldwide spread of the epidemic is an important research topic. Effectively predicting the daily number of newly confirmed cases of COVID-19 is of great significance to the formulation of epidemic prevention and control policies and the development of economic and social activities during the entire stage of the epidemic, especially to provide important forecast information for the allocation of medical resources and policy formulation during the outbreak.
Fig. 1.
New daily confirmed cases of COVID-19.
Fig. 2.
New daily death cases in world.
Now, there are three main types of prediction methods for COVID-19: 1. Traditional infectious disease model; for example, Jia et al. [2] used a dynamic expansion susceptibility clearance model(eSIR) of infectious diseases with different intervention effects in different periods to estimate the epidemic trend in Italy. Bastos et al. [3] used the SIR model with added parameters to model and predict the evolution of the Brazilian COVID-19 pandemic. Yang et al. [4] used population migration data and the latest COVID-19 epidemiological data to integrate into the SEIR model to obtain the epidemic curve. 2. Machine learning model; for example, Tomara and Guptab [5] used data-driven estimation methods such as long-term memory (LSTM) and curve fitting to predict the number of cases of COVID-19 in India and the impact of preventive measures such as social isolation and blockade on the transmission of COVID-19 in India. Hu et al. [6] developed an improved stacked automatic encoder and predicted the cumulative confirmed case curve in China. Sina et al. [7] compared and analyzed machine learning model and soft computing model to predict the outbreak of COVID-19. 3. Time series model; for example, Petropoulos and Makridakis [8] used the exponential smoothing model to predict the confirmed cases of COVID-19, and the results showed that the number of confirmed COVID-19 cases would continue to increase. Benvenuto et al. [9] used the autoregressive integrated moving average model (ARIMA) to predict the epidemiological trend of COVID-19 prevalence and incidence rate in 2019. Maleki et al. [10] proposed a regression time series model based on two scale mixed normal distribution to analyze the real time series data of confirmed and recovered COVID-19 cases. All the above prediction models can effectively fit and predict the development of COVID-19 epidemic.
However, the epidemic model is hypothetical and may be affected by factors such as geography and super communicators [11]. From the above three types of models, it can be seen that the infectious disease model and time series model may need a large amount of data for accurate parameter identification. Literatures [12], [13], [14] point that the machine learning model may need a large amount of data for training and testing to achieve accurate results. In the research on the development trend of infectious diseases, Richards model [15] has been widely used in a variety of infectious diseases due to its advantages in processing saturated S-shaped data. Hsieh [16] used Richards model to fit the number of SARS cases in many places in China, and estimated the parameters and the maximum number of cases to illustrate the turning point. Hsieh and Ma [17] matched the Richards model with dengue fever notification numbers to detect turning points in the epidemic. Chan et al. [18] used Richards model to estimate the basic reproductive number (R0) of cholera and the proportion of unrecognized cases. The heterogeneity of R0 estimates generated by the model was consistent with the dynamic changes of cholera described. Wang et al. [19] started from a simple epidemic SIR model and re-examined the Richards model through the internal connection between the two models. More accurate and stable model parameters and key epidemic characteristics were estimated in H1N1, SARS and other epidemics. The accurate parameter identification of this model is also inseparable from a large-number data.
For the new type of coronavirus such as COVID-19, due to the low availability of case data and incomplete knowledge, especially in the early stage of the outbreak, the information is incomplete and the amount of data is small. The above models may not have obvious effect on the data of uncertainty phenomenon. Therefore, it is considered to be attractive to find a prediction model with less information to obtain relatively effective results [20]. In recent years, in systems that deal with incomplete information, the grey model stands out by virtue of its "simple model, strong adaptability, and easy parameter changes". It is widely used in energy, finance, transportation, environment, manufacturing, materials and other industries [[12], [13], [14],[20], [21], [22], [23], [24], [25]]. In the field of infectious diseases, the grey model is also widely used. Guo et al. [26] used traditional GM(1,1) and SMGM(1,1) based on self-memory principle to predict the incidence of three typical infectious diseases in China. Wang et al. [27] used GM(1,1) for prediction of hepatitis B in China. Zhang et al. [28] used GM(1,1), the grey period extended combined model, and the improved Fourier series grey model to predict Hydatid disease, and these models successfully predicted the development of these diseases. The above models are all based on the GM(1,1) model. The GM(1,1) model is suitable for strong exponential growth data and is a linear model. The research object of this article is the fluctuating S-shaped data, so the applicability of GM(1,1) is limited. Therefore, this paper chooses the nonlinear grey model as a research method to predict the daily number of newly confirmed cases of COVID-19 and provide an important basis for formulating epidemic prevention policies.
In the grey system, the Verhulst model has strong prediction ability for single-peak or saturated S-shaped sequence [29]. Wang et al. [11] used the rolling Verhulst model to predict the final number of COVID-19 infection cases, and good results were achieved. Şahin and Şahin [30] used fractional Nonlinear Grey Bernoulli model to predict the cumulative number of cases in Italy, the United Kingdom and the United States. The Verhulst model is a special form of the grey Bernoulli model, and the whitening equation of the Verhulst model is the logistic model. The Richards model is also a generalized logistic model [15]. Because of its good performance on S-shaped data and its relationship with the Verhulst model, this paper study the corresponding grey prediction model based on the Richards model.
Therefore, based on the characteristics of COVID-19 and the research status of Richards model and grey prediction model, this paper uses the difference information principles of the grey prediction model to derive the grey Richards prediction model on the basis of the Richards model, and the grey covid-19 prediction model (GERM(1,1,)) is established with the help of the effect of grey action quantity. Using the relevant grey knowledge to solve the model, the time response function of the model is obtained, and the properties of the optimization model are studied. Through practical cases, the results of the new model are compared with the grey linear model, the grey nonlinear model and the autoregressive model, so as to verify the effectiveness of the model and provide important information for the government to formulate economic policies.
So, the main contributions of this paper are as follows:
-
1
Richards model is widely used in many kinds of infectious diseases. Based on the structure of traditional Richards growth model and the differential information principle, namely the relationship between differential equation and difference equation, the traditional Richards growth model is transformed into corresponding grey prediction model.
-
2
Due to the limitation of the traditional model for data trend, the new model uses natural index to improve the grey action of the grey Richards model, and optimizes the nonlinear term and background value of the model by GA algorithm, so as to weaken or even eliminate the dependence of the traditional Richards model on saturated S-shaped data, and improve the accuracy and applicability of the traditional model, which is also a generalization of Richards model and grey prediction model.
-
3
Through the empirical analysis of small-sample and large-sample data, it shows that the new model can effectively carry out short-term and medium-term prediction and makes up for the defects of grey prediction model generally used for short-term prediction. The new model also effectively predicts the daily number of newly confirmed COVID-19 cases in four countries, which will provide help for local governments to make policy decisions.
The rest of this paper is as follows: Section 2 establishes the GERM(1,1,) model, and studies the properties and mechanism of the new model; Section 3 analyses the validity of the new model; Section 4 discusses the application of the GERM(1,1,) model. Section 5 presents the conclusions.
In the full text, the different abbreviations are for different grey prediction models. Abbreviations and their meanings are listed in Table 1 .
Table 1.
Abbreviations of models.
| Number | Abbreviation | Definition |
|---|---|---|
| 1. | GM(1, 1) | Grey model with one variable and one first order equation [31] |
| 2. | Verhulst | Verhulst grey model [32] |
| 3. | ARGM(1,1) | Autoregressive grey model [33] |
| 4. | ONGM(1,1) | Optimized NGM(1,1,k,c) model [34] |
| 5. | ENGM(1,1) | Exact nonhomogeneous grey model [35] |
| 6. | ARIMA | Autoregressive Integrated Moving Average model [36] |
| 7. | NGBM(1,1) | Nonlinear Grey Bernoulli Model [37] |
| 8. | GRM(1,1) | Grey Richards model |
| 9. | GERM(1,1,) | Grey Extend Richards model |
2. Theoretical modeling of GERM(1,1,)
This section first introduces the related concepts and properties of Richards model, then establishes the corresponding grey prediction model according to the grey difference information and grey action quantity, and finally optimizes the parameters of the new model by GA algorithm.
2.1. Classical growth Richards model
The logistic model was first proposed by Verhulst [38] in 1838 to simulate population growth after Malthus model [39]. The model equation, also known as Verhulst equation, is as follows:
| (1) |
Where is the population size in question at time , is the intrinsic growth rate, and is maximum capacity.
In 1959, Richards [15] proposed the following modification of the logistic model to model growth of biological populations:
| (2) |
where, is the cumulative number of infection cases at time , is the carrying capacity of the outbreak or the total number of cases, is the per capita growth rate of the infected population, parameter provides the flexibility measure of S-shaped curvature shown by the result solution curve, that is, the degree of deviation from the S-shape dynamics of the classical logistic growth model. When , Richards model becomes the logistic model. Richards model is used to predict the spread of diseases. It only considers the cumulative population size with saturated growth as the model and dynamics of infectious disease outbreak and development, which is due to the reduction of cases due to the attempt to avoid contact and the implementation of control measures.
2.2. Description of the GRM(1,1)model
Let the daily number of newly confirmed cases be
| (3) |
The first- accumulating generation operator(1-AGO) sequence is
| (4) |
where
Eq. (2) is a first-order nonlinear differential equation, which is continuously differentiable and contains infinite information. However, the number of confirmed cases per unit time is only a discrete time series with limited uncertain information or so-called grey information. Therefore, Eq. (2) should be transformed and discretized according to the principle of differential information, so as to adapt to the characteristics of grey information [40]. This principle is also an important cornerstone of grey model [31]. It is systematically and accurately described in monograph [41].
Therefore, the grey Richards model (GRM(1,1)) is introduced as below. In Eq. (2), represents the number of all the patients diagnosed in unit time, that is, the total number of all patients diagnosed in a certain period of time. According to different unit time, it can be the number of patients diagnosed per minute, the number of patients diagnosed per hour, the number of patients diagnosed daily, and the number of patients diagnosed per week. If the initial time point is recorded as , then the number of confirmed patients is recorded times between the time periods , and the number of all patients diagnosed is
| (5) |
is a 1-AGO sequence of Eq. (4), which is similar to that of oil production in a certain period of time in [42,43] and energy consumption in a certain period in [44]. Therefore, according to Eq. (2), the following formula holds
| (6) |
Substituting the first-order difference for the differential at the left end of Eq. (6), then at :
| (7) |
Thus, the following grey prediction models can be defined:
Definition 1
Set and as Eqs. (3) and (4), then the sequence is called the nearest mean generating sequence of :
(8)
where .
It should be noted that is usually regarded as a grey derivative and can provide the necessary information related to the time-dependent function . That is, can be regarded as the grey derivative sequence of and replace during . In addition, the background value of can be formalized as
| (9) |
Definition 2
Set , and as Eqs. (3), (4) and (8), so,
(10)
is grey Richards model (GRM(1,1)). Set , Eq. (10) becomes
| (11) |
Eq. (11) is also a power model in grey model.
Since the daily data of new patients of COVID-19 shows a fluctuating S-shaped trend, and Richard model is suitable for processing data close to single-peak and saturated S-shaped [15], [16], [17], [18], [19], this paper uses a new grey action to improve structure of the grey Richard model, it is suitable for the prediction of daily number of confirmed cases of COVID-19.
2.3. Description of the GERM(1,1,) model
Obviously, the optimization of grey action is an effective mean to improve the performance and applicability of grey model [45]. In this section, a new driving grey model is proposed, which takes the natural exponential function of time as the grey action quantity. Therefore, this section gives the definition of the grey extended Richards model, the time response function and the restored values of the model.
Definition 3
Based on the Sections 2.1 and 2.2, the Grey Extend Richards model (GERM(1,1,)) is defined as
(12)
where is the development coefficient, the term denotes the power-driven grey input. and are nonlinear quantities which can be tunable.
If , the GERM(1,1,) becomes GM(1,1,) [45]. Set , Eq. (12) becomes
| (13) |
The solution of Eq. (13) is
| (14) |
So, based on the , the time response function can be derived from Eq. (14) by
| (15) |
where , and the restored value is
| (16) |
Definition 4
Assume and are defined as the same in Definition 1. The background value is optimized by extrapolation background value [23], is defined as
(17)
So, the parameters of GERM(1,1,) are computed by the following expression
| (18) |
where
Proof
By integrating on both side of Eq. (12), it becomes
(19)
It follows from Eq. (19) that
| (20) |
Using the trapezoidal formula, the terms and can be computed by
| (21) |
| (22) |
By substituting Eqs. (21) and (22) into Eq. (20), Eq. (20) becomes
| (23) |
Once are given, the linear parameters can be straightforwardly computed by the following expression
where
Therefore Definition 4 is proven.
2.4. Error evaluation criteria
This subsection provides some standard error evaluation criteria to measure the accuracy of grey forecasting models. In general, they are defined in Table 2 . Meanwhile, Lewis’ criterion [46] shown in Table 3 is used to illustrate the prediction ability of the model.
Table 2.
Metrics for evaluating effectiveness of the models.
| Name | Abbreviation | Formulation |
|---|---|---|
| The absolute percentage error | APE | |
| The mean absolute simulation percentage error | MAPESIM | |
| The mean absolute prediction percentage error | MAPEPRE | |
| The total mean absolute percentage error | MAPETOT | |
| Root mean squares percentage error | RMSPE | |
| Mean absolute percentage error | MAE | |
| Index of agreement | IA | |
| Theil U statistic 1 | U1 | |
| Theil U statistic 2 | U2 | |
| Correlation coefficient | R |
Table 3.
Lewis’ criterion for model evaluation.
| MAPE (%) | Prediction performance |
|---|---|
| <10 | Excellent |
| 10-20 | Good |
| 20-50 | Reasonable |
| >50 | Incorrect |
2.5. Optimization of nonlinear parameters
It can be seen from Section 2.4 that the parameters of GERM(1,1,) can be obtained through the values of three parameters , where are nonlinear quantities. In this section, the genetic algorithm (GA) is used to search for , and MAPETOT is taken as the objective function. Then, the mathematical expression of the optimization problem is
| (24) |
GA is proposed by American Professor John Holland [47]. It randomly searches the optimal value by simulating the genetic law and evolutionary theory of natural organisms. It applies the evolutionary principle of survival of the fittest and elimination of the unfit in the biological world to optimize the parameter individuals after coding. Through selection, variation and crossover, the overall fitness level of the population is improved continuously, which not only inherits the good information of the previous generation, but also is superior to the previous generation. This is repeated until the desired conditions are met. The GA algorithm is utilized to search for optimal , and its main procedures are given in Algorithm 1 .
Algorithm 1.
The GA algorithm to find the optimal .
| Set the objective function and the maximum iteration number Input: The original parameters , original data and the number of modelling data Output: The best for do Substitute to and obtain parameters Substitute parameters to discrete equation Eq. (14) and compute the simulation value to obtain Compute in Eq. (16) Compute APE and MAPE in Table 2. End Update the minimum MAPE value Return the best by the GA algorithm. |
3. Validation of the GERM(1,1,) model
This section mainly discusses the effectiveness of the new model through two data sets. The two selected data sets are divided into slowly growing small-sample data and large-sample data to illustrate that the new model has improved the accuracy of GRM(1,1) after modifying the grey action quantity, and the dependence of GRM(1,1) on saturated S-shaped data is weakened. Therefore, the traditional GM(1,1) and GRM(1,1) are selected for comparison. GRM(1,1) is also the form of NGBM(1,1). The validity comparison of the models can be explained from two aspects: 1. Using eight evaluation indexes in Table 2, the smaller APE, MAPE, RMSPE, MAE, MSE, U1 and U2, the higher the accuracy of the model, and vice versa. The higher the values of IA and R, the higher the accuracy of the model. 2. Use the comparison chart of APE value every year.
3.1. Validation case 1: fitting of small-sample data
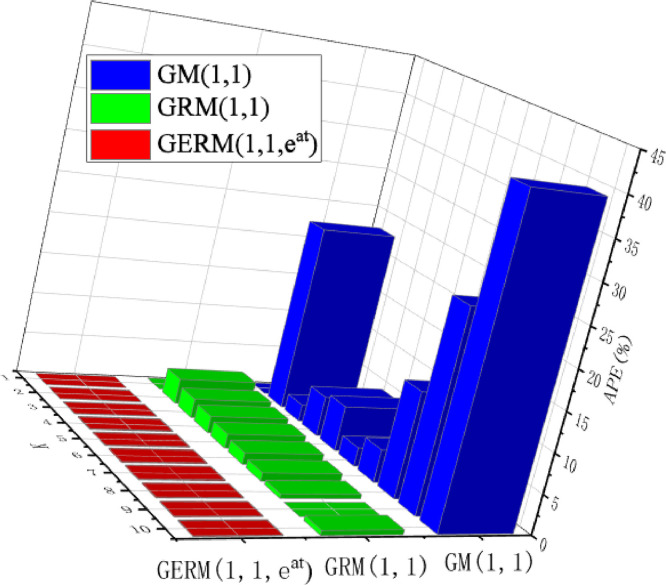
The first case study object is small sample data. The first seven data are used for modeling, and the last three data are used to test the model accuracy. According to GA algorithm, the optimal parameter of GRM(1,1) is , the optimal parameters of GERM(1,1,) are . The calculation results of three grey models are shown in Table 4 and the index results are shown in Table 5 . In Table 4, the MAPESIM and MAPEPRE of GM(1,1) are the highest. In the modeling phase, MAPESIM of GERM(1,1,) is nearly 2% higher than that of GRM(1,1), and the prediction error is the smallest. In Table 5, the MAPETOT of GERM(1,1,) is 1.3% higher than that of GRM(1,1). In addition, the other indicators of GERM(1,1,) are good, and the R index reaches 1. In order to further show the fitting effect of the model, the APE value comparison chart is made, as shown in Fig. 3 . In Fig. 3, the APE values of GERM(1,1,) are basically the lowest, and compared with the other two models, the APE values are almost zero every year. The comparison results of the above two aspects show that the GERM(1,1,) improves the accuracy of the original GRM(1,1).
Table 4.
Results of three grey models in validation Case 1.
| Raw data | GM(1,1) model | APE (%) | GRM (1,1) | APE (%) | GERM (1,1,) | APE (%) |
|---|---|---|---|---|---|---|
| 1 | 1.0000 | 0.0000 | 1.0000 | 0.0000 | 1.0000 | 0.0000 |
| 2 | 2.4658 | 23.2912 | 1.9385 | -3.0731 | 1.9994 | -0.0312 |
| 3 | 3.0619 | 2.0644 | 2.9302 | -2.3283 | 3.0019 | 0.0633 |
| 4 | 3.8021 | -4.9464 | 3.9194 | -2.0157 | 4.0041 | 0.1025 |
| 5 | 4.7213 | -5.5739 | 4.9133 | -1.7336 | 5.0061 | 0.1201 |
| 6 | 5.8627 | -2.2889 | 5.9161 | -1.3993 | 6.0075 | 0.125 |
| 7 | 7.2802 | 3.9993 | 6.9302 | -0.9970 | 7.0088 | 0.1257 |
| MAPESIM(%) | 7.0273 | 1.9245 | 0.0944 | |||
| 8 | 9.0399 | 12.9983 | 7.9578 | -0.528 | 8.0097 | 0.1213 |
| 9 | 11.2252 | 24.7247 | 9.0002 | 0.0021 | 9.0103 | 0.1144 |
| 10 | 13.9389 | 39.3890 | 10.0587 | 0.5874 | 10.0106 | 0.1061 |
| MAPEPRE(%) | 25.704 | 0.3725 | 0.1139 |
Table 5.
Metrics of models in Validation Case 1.
| Metrics | GM (1,1) | GRM (1,1) | GERM (1,1,) | GERM (1,1,) rank |
|---|---|---|---|---|
| MAPETOT(%) | 19.8793 | 1.4072 | 0.1009 | 1 |
| RMSPE | 17.1908 | 1.5973 | 0.1001 | 1 |
| MAE | 0.8626 | 0.0554 | 0.0060 | 1 |
| MSE | 2.1983 | 0.0040 | 5.03E-05 | 1 |
| IA | 0.95181 | 0.99988 | 0.999998 | 1 |
| U1 | 0.1093 | 0.0051 | 0.0005 | 1 |
| U2 | 0.2390 | 0.0102 | 0.001143 | 1 |
| R | 0.9749 | 0.9999 | 1.0000 | 1 |
Fig. 3.
APE of the three models in Validation Case 1.
3.2. Validation case 1: fitting of large-sample data
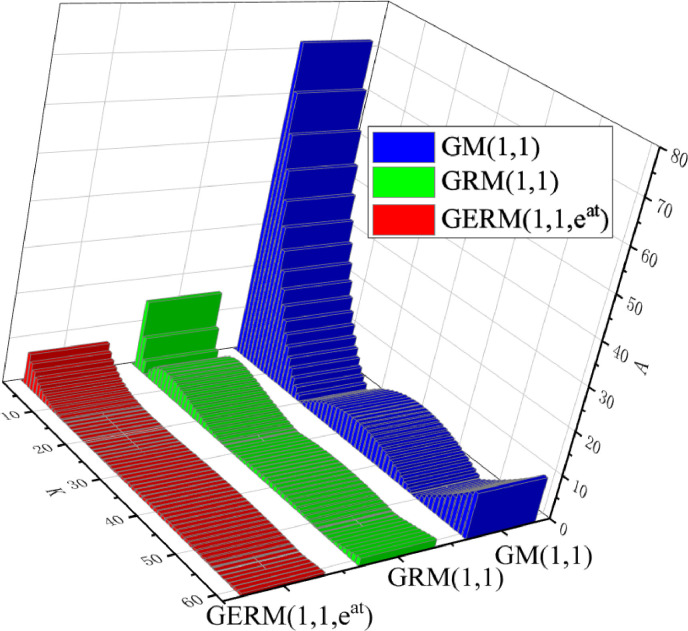
The first case study object is small sample data. The first seven data are used for modeling, and the last three data are used to test the model accuracy. The optimal parameter of GRM(1,1) is , the optimal parameters of GERM(1,1,) found by GA algorithm are . Due to the large amount of data, the original data and the calculation results of the three grey models are shown in Table 12 in Appendix A, and the index results are shown in Table 6 . In Table 6, the MAPETOT of GERM(1,1,) is the lowest, which is 1 percentage point higher than the original GRM(1,1), and the results of other seven indicators are the best. Meanwhile, the R of this cases is also 1. As in the previous case, the APE comparison chart between the models are drawn, as shown in Fig. 4 . In Fig. 4, the APE values of the GERM(1,1,) are basically the lowest, which is closer to 0 from the 11th point compared with the other two models. In conclusion, GERM(1,1,) improves the accuracy of GRM(1,1) model.
Table 6.
Metrics of models in Validation Case 1.
| Metrics | GM (1,1) | GRM (1,1) | GERM (1,1,) | GERM (1,1,) rank |
|---|---|---|---|---|
| MAPETOT(%) | 10.2098 | 1.7895 | 0.7782 | 1 |
| RMSPE | 17.1908 | 1.5973 | 0.1001 | 1 |
| MAE | 0.8626 | 0.0554 | 0.0060 | 1 |
| MSE | 2.1983 | 0.0040 | 5.03E-05 | 1 |
| IA | 0.9518 | 0.99988 | 0.999998 | 1 |
| U1 | 0.1093 | 0.0051 | 0.0005 | 1 |
| U2 | 0.2390 | 0.0102 | 0.001143 | 1 |
| R | 0.9749 | 0.9999 | 1.0000 | 1 |
Fig. 4.
APE of the three models in Validation Case 2.
3.3. Analysis of result
In two practical cases, MATLAB software is used to calculate the whole simulation process. According to the fitting results of three grey models and APE comparison chart, the following conclusions can be obtained:
-
(1)
In the contrast test with the eight metrics of traditional GM(1,1) and GRM(1,1) (NGBM(1,1)), those of GERM(1,1,) are the best. Meanwhile, the APE value of GERM(1,1,) are basically the lowest, that is to say, its error is the smallest and the accuracy is the highest, indicating that the GERM(1,1,) is competitive.
-
(2)
From original GRM(1,1) to GERM(1,1,), the results of two cases show that the accuracy of the extended model is significantly improved. It shows that the extended grey action model can improve the structure of the grey model, alleviate the exponential growth and saturated S-shaped growth, weaken the dependence of the original GRM(1,1) on the saturated S-shaped data, and overcome the disadvantage of the grey model used in short-term prediction, and apply new model to medium and long-term prediction.
4. Applications
After the model test in Section 3, GERM(1,1,) is applied to the representative epidemic developing countries in the world, including China, Italy, the United Kingdom and Russia. The research data on the number of all confirmed patients per day are all from the WHO [1]. The first three cases use small-sample data, and the last case uses large-sample data. In addition to the traditional GM(1,1), GRM(1,1) (NGBM(1,1)), Verhulst model, ARGM(1,1), ENGM(1,1), ONGM(1,1) and ARIMA models are added to the model for comparison. There are still two ways to compare: one is to use MAPE index; the other is curve trend chart and APE percentage chart. The curve trend chart is used to evaluate the fitting and approximation degree of the model simulation trend line and the actual data trend line. The higher the degree of fitting and approximation, the better the data fitting ability of the model is. The APE percentage comparison chart is used to evaluate the error size by measuring the area occupied by percentage of APE value of each point of each model. The larger the area proportion, the greater the error. The number of modeling data and prediction data in the four cases is shown in Table 7 . Due to the large amount of data in each case, the fitting result of each model is put into the Appendix.
Case 1: China
Table 7.
Metrics of models in Validation Case 1.
| NO. | Country | Date | Points for modeling | Points for prediction |
|---|---|---|---|---|
| Case 1 | China | 1.23-2.6 | 9 | 6 |
| Case 2 | Italy | 3.10-3.21 | 10 | 2 |
| Case 3 | United Kingdom | 4.11-4.25 | 5 | 10 |
| Case 4 | Russian | 6.1-8.12 | 61 | 12 |
China is the first center for the occurrence of COVID-19 epidemic, and China immediately adopted strong measures to quickly begin to control the epidemic. In this case, eight prediction models are established based on the number of confirmed cases in the first nine days of the outbreak period, and the data of the next six days are used to test the accuracy of the models. The fitting results of the eight prediction models are shown in Table 13 in Appendix, and the comparison of MAPE values is shown in Table 8 . Using GA algorithm, the optimal parameters of GERM(1,1,) are , and the optimal parameter of GRM (1,1) is .
Table 8.
Fitting MAPE values of models in Case 1.
| MAPE | GM(1,1) | Verhulst | ARGM(1,1) | ONGM(1,1) | ENGM(1,1) | ARIMA | GRM(1,1) | GERM(1,1,) |
|---|---|---|---|---|---|---|---|---|
| MAPESIM | 29.7152 | 43.2128 | 35.3292 | 14.6649 | 17.5457 | 72.7097 | 24.7328 | 7.7121 |
| MAPEPRE | 59.5837 | 55.2802 | 27.2007 | 12.1904 | 14.1994 | 10.0138 | 5.2691 | 4.2684 |
| MAPETOT | 42.5160 | 48.3845 | 31.8456 | 13.6044 | 26.7355 | 53.4474 | 16.3912 | 6.2362 |
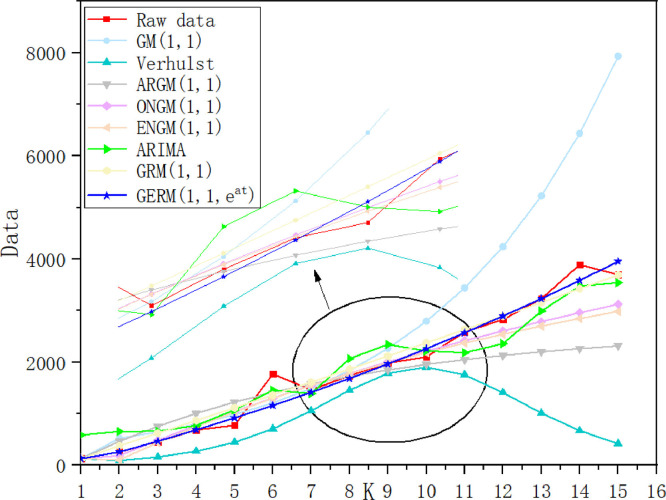
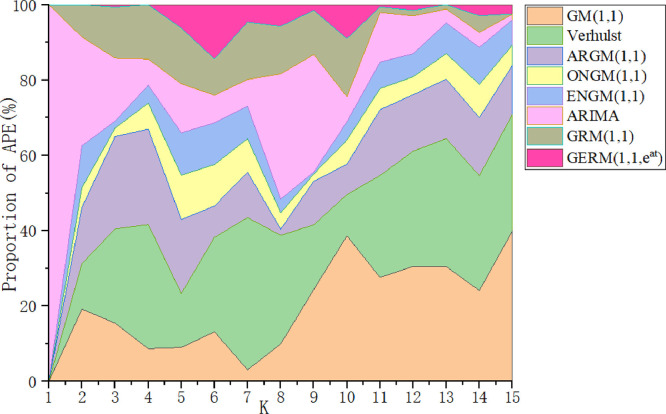
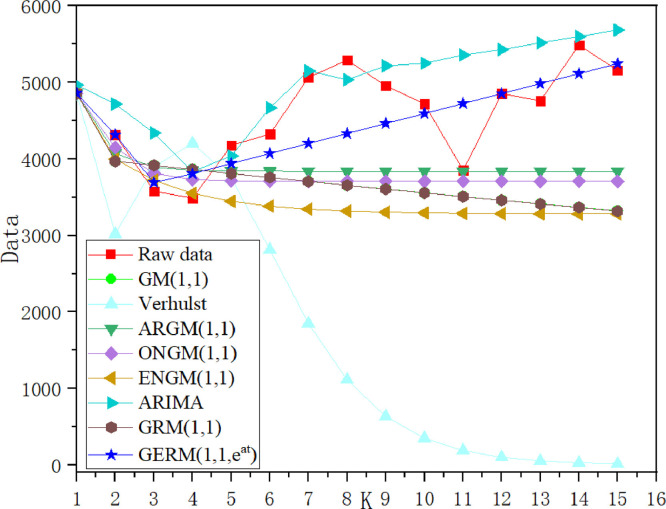
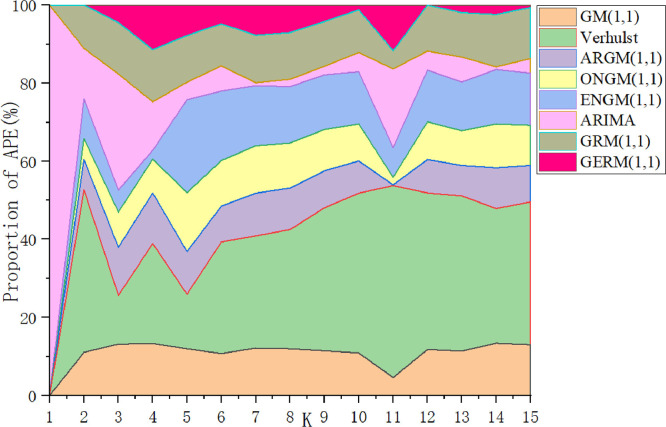
In Table 8, in the modeling stage, the MAPESIM of GERM(1,1,) is the lowest, which is 17% higher than that of the original GRM(1,1). The two models with the largest MAPESIM are ARIMA and Verhulst models. In the prediction stage, the lowest MAPEPRE is GERM(1,1,), the two models with the maximum MAPEPRE are also ARIMA and Verhulst models, which indicates that ARIMA and Verhulst models are not suitable for the prediction of epidemic situation in China. In order to show the model fitting effect from the second aspect, the results in Table 13 are transformed into curve trend chart and APE percentage chart, as shown in Fig. 5, Fig. 6 . In Fig. 5, the GERM(1,1,) is the closest to the original trend line. The original GRM(1,1) basically overestimates the development of the epidemic situation, first overestimates and then underestimates the original data, Verhulst shows a single-peak trend, ENGM(1,1) and ONGM(1,1) basically underestimate the development of the epidemic situation. In Fig. 6, it is clear that ONGM (1,1) and ENGM (1,1) occupy a small area in the percentage of APE each year, indicating that the prediction error of these two models are small, while the APE percentage of GERM(1,1,) is slightly larger at point 6 and point 10, and the area of other points is basically close to 0. The comparison results of the two aspects show that GERM(1,1,) effectively predicts the development of the early stage of the outbreak in China.
Case 2: Italy
Fig. 5.
the overall trend of simulation results of eight models in Case 1.
Fig. 6.
APE percentages of the eight models in Case 1.
At the beginning of the outbreak in Italy, the number of newly COVID-19 confirmed cases in a single day is close to 3500, and its mortality rate exceeded 7%. Two data made Italy an early outbreak country in Europe. If calculated in proportion to the population, the epidemic in Italy is the most severely affected country in the world at that time. The data of 10 days from March 10 to March 19 are used to build eight models, and the data from the next two days are used to test the model prediction accuracy. According to GA algorithm, the optimal parameters of GERM(1,1,) are , and the optimal parameter of GRM(1,1) is . The fitting results of the eight prediction models are shown in Table 14 in Appendix, and the comparison of MAPE values is shown in Table 9 .
Table 9.
Fitting MAPE values of models in Case 2.
| MAPE | GM(1,1) | Verhulst | ARGM(1,1) | ONGM(1,1) | ENGM(1,1) | ARIMA | GRM(1,1) | GERM(1,1,) |
|---|---|---|---|---|---|---|---|---|
| MAPESIM | 8.3361 | 52.5502 | 14.7858 | 8.3953 | 19.0126 | 21.3977 | 8.3376 | 7.1304 |
| MAPEPRE | 11.7031 | 29.7941 | 21.7826 | 5.8776 | 21.9019 | 24.2912 | 11.7076 | 3.4857 |
| MAPETOT | 8.9483 | 48.4127 | 16.0579 | 7.9375 | 19.5379 | 21.9238 | 8.9503 | 6.4677 |
In Table 9, in the modeling stage, the MAPESIM of GERM(1,1,) is the lowest, which is 1% higher than that of GRM(1,1). In the prediction stage, the MAPEPRE of GERM(1,1,) is the lowest, which is 8% higher than that of GRM(1,1), so the MAPETOT of GERM(1,1,) is also the lowest. The results in Table 14 are transformed into curve trend chart and APE percentage chart, as shown in Fig. 7, Fig. 8 . In Fig. 7, the blue line is the closest to red line, that is, the line of GERM(1,1,) model is the closest to the original trend line. GRM(1,1), Verhulst, ARIMA, ENGM(1,1) and ONGM(1,1) basically overestimate the development of the epidemic. In Fig. 8, the APE of Verhulst model occupies the largest area. APE of GERM(1,1,) only has larger areas at points 5, 6, and 10 than other points, while other points are basically close to 0. The above results show that GERM(1,1,) effectively predict the development in the early stage of the outbreak in Italy.
Case 3: The United Kingdom
Fig. 7.
The overall trend of simulation results of eight models in Case 2.
Fig. 8.
APE percentages of the eight models in Case 2.
After the outbreak in Italy, the epidemic began to spread in Europe, and the United Kingdom was not included. The research objects of the first two cases were modeling data with a large amount of modeling data and a small amount of predicted data. Therefore, this case selects the data of 5 days from April 11 to April 15 during the outbreak period of the United Kingdom to establish the model, and the data of the last 10 days are used for model accuracy test. Based on GA algorithm, the optimal parameters of GERM(1,1,) are , and the optimal parameter of GRM(1,1) is . The fitting results of the eight prediction models are shown in Table 15 in Appendix, and the comparison of MAPE values is shown in Table 10 .
Table 10.
Fitting MAPE values of models in Case 3.
| MAPE | GM(1,1) | Verhulst | ARGM(1,1) | ONGM(1,1) | ENGM(1,1) | ARIMA | GRM(1,1) | GERM(1,1,) |
|---|---|---|---|---|---|---|---|---|
| MAPESIM | 9.2207 | 17.4099 | 8.1942 | 7.1084 | 7.6501 | 9.2277 | 9.2222 | 4.4999 |
| MAPEPRE | 26.2797 | 84.8051 | 20.0258 | 22.7735 | 31.1187 | 10.9995 | 26.3003 | 8.9448 |
| MAPETOT | 21.4057 | 21.4208 | 16.6453 | 18.2976 | 24.4134 | 10.4933 | 21.4208 | 7.6748 |
In Table 10, in the modeling stage, the MAPESIM of GERM(1,1,) is the lowest, which increases that of GRM(1,1) by 5 percentage points, and the highest MAPESIM model is Verhulst. In the prediction stage, the MAPEPRE of GERM(1,1,) is the lowest, which increases that of GRM(1,1) model by 18 percentage points. The MAPESIM of ARIMA model is the second, and MAPESIM of other models is more than 20%. Therefore, the MAPETOT of GERM(1,1,) is also the lowest. The results in Table 15 are transformed into curve trend chart and APE percentage chart, as shown in Fig. 9, Fig. 10 . In Fig. 9, the line of GERM(1,1,) is the closest to the original trend line. The trend line of ARIMA model is similar to the actual trend line, but some points are far away from the actual point. The other six models basically underestimate the development of epidemic situation. In Fig. 10, GM (1,1), ARIMA and Verhulst models have larger area of APE percentage in modeling stage, the APE percentage of ARIMA and Verhulst models are also larger in prediction stage, and APE percentage of GERM(1,1,) only accounts for a large area in points 4, 7 and 11, and these of other points are close to 0. The above results show that GERM(1,1,) can effectively predict the development of the outbreak period in Britain.
Case 4: Russian
Fig. 9.
The overall trend of simulation results of eight models in Case 3.
Fig. 10.
APE percentages of the eight models in Case 3.
Up to now, the daily diagnosis of more than 5000 patients in Russia, the epidemic situation is still grim. The research objects of above three cases are small-sample data, and the last case uses large-sample data. The data of June and July are used for modeling, and the data of August are used to test the model accuracy. Base on the GA algorithm, the optimal parameters of GERM(1,1,) are , and the optimal parameter of GRM(1,1) is . The comparison of MAPE values is shown in Table 11 , and the fitting results of the eight prediction models are shown in Table 16 in Appendix.
Table 11.
Metrics of models in Case 4.
| MAPE | GM(1,1) | Verhulst | ARGM(1,1) | ONGM(1,1) | ENGM(1,1) | ARIMA | GRM(1,1) | GERM(1,1,) |
|---|---|---|---|---|---|---|---|---|
| MAPESIM | 2.8280 | 51.8554 | 3.0286 | 2.6521 | 2.6664 | 1.4243 | 2.8294 | 2.6248 |
| MAPEPRE | 2.7481 | 40.7817 | 3.9221 | 1.0399 | 1.8335 | 7.0740 | 2.7735 | 0.9023 |
| MAPETOT | 2.8149 | 50.0351 | 3.1755 | 2.3871 | 2.5295 | 2.3530 | 2.8202 | 2.3416 |
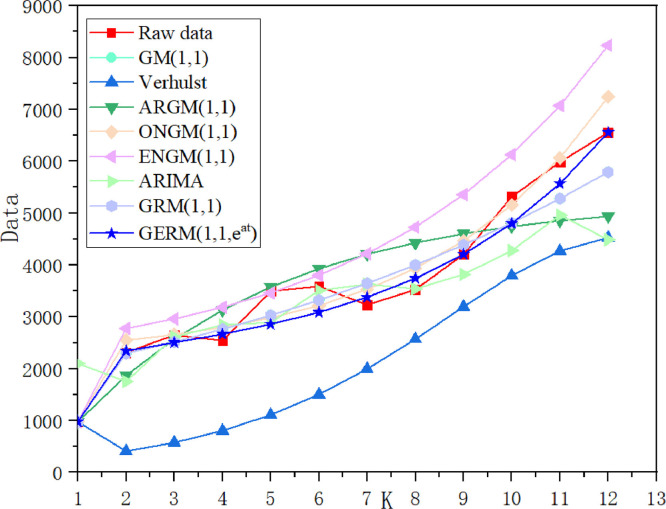
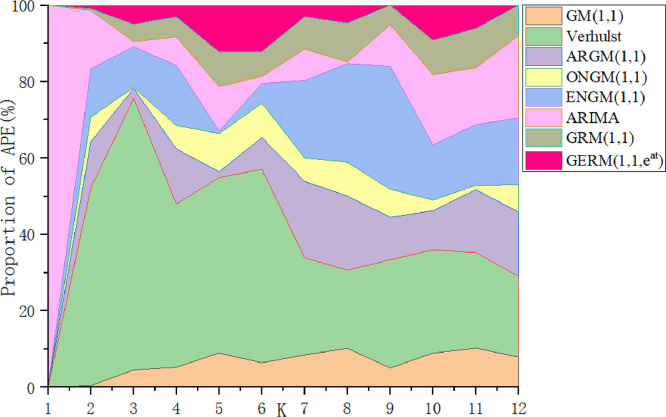
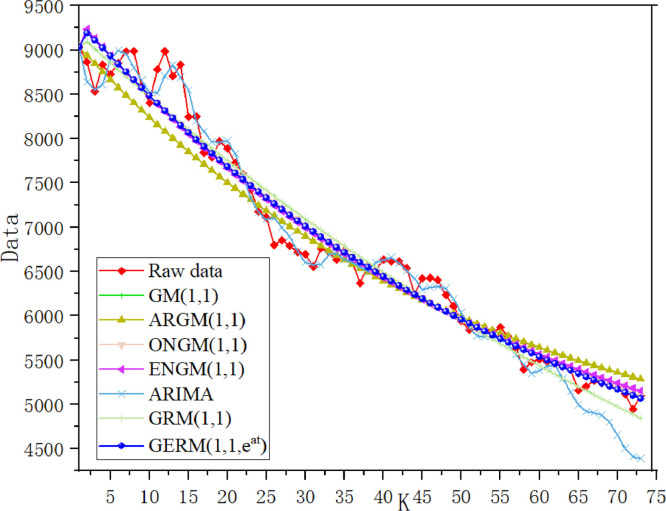
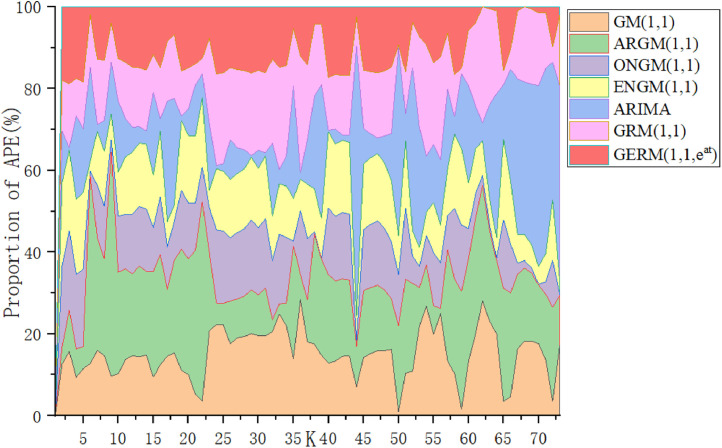
Table 11 summarizes the errors in the modeling and prediction stages. In the modeling phase, the ARIMA model has the lowest MAPESIM, followed by the GERM(1,1,), and MAPESIM of Verhulst model is the largest. In the prediction phase, the MAPEPRE of GERM(1,1,) is the best, which is about 2% higher than that of original GRM(1,1). Due to the large error of Verhulst model, the graph of Verhulst model is omitted in the process of graphical fitting results. In Fig. 11 , ARIMA model has the best fitting effect in the modeling stage, while the predicted fitting line of GERM(1,1,) is the closest to the actual curve, which indicates that ARIMA model is more suitable for fitting large-sample data. In Fig. 12 , in the modeling phase, the APE percentage of ARIMA model accounts for the smallest area, and other models have little difference. In the prediction phase, only two points of GERM(1,1,) account for a large proportion, and other points are close to 0. The comparison results of the two perspectives show that the expanded model not only can effectively predict the daily number of confirmed patients in Russia, but also has advantages in medium and long-term prediction, which improves the disadvantage of grey model which is generally suitable for short-term prediction.
Fig. 11.
the overall trend of simulation results of eight models in Case 4.
Fig. 12.
APE percentages of the eight models in Case 4.
5. Conclusion
Due to the advantages of traditional Richards model in processing saturated S-shaped data, it has been applied to the trend prediction of various infectious diseases. Therefore, in this paper, based on the structure of traditional Richards growth model and the theory of grey differential information, the corresponding GRM(1,1) is established. Meanwhile, the grey action quantity is modified to improve the structure of GRM(1,1), and the GERM(1,1,) is established. The new model weakens the dependence of the Richards model on S-shaped data. In practical cases, through the comparison with GM(1,1), ARGM(1,1), ONGM(1,1), ENGM(1,1), Verhulst and ARIMA models, GRM(1,1) (NGBM(1,1)), the fitting effect of GERM(1,1,) is better than other models, that is, this model accuracy is the highest.
As mentioned in this paper, China, Italy, Britain, Russia and other places are the regions with severe epidemic situation in different periods of time, and the prediction of their infectious disease system is more complex. The priority of the GERM(1,1,) over the other above models indicates that it is qualified to predict the number of daily confirmed patients of COVID-19. As COVID-19 is in a global pandemic, it is expected that the GERM(1,1,) model can be applied to predict the number of confirmed cases around the world, which will be of great help to local governments to make policy decisions
As a single-variable grey model, GERM(1,1,) can effectively predict the daily average number of daily confirmed patients of COVID-19. With the in-depth study of covid-19, the mechanism of virus transmission and the factors affecting virus transmission will become clear gradually. In the process of prediction, considering these factors such as temperature, population and environment will bring uncertainty to the model. Therefore, fully mining the ways of these factors affecting virus transmission, and introducing them into the GERM(1,1,), and extending the GERM(1,1,) to a multivariate model, the prediction effect may be further improved. How to establish multivariate GERM model is also our anticipated next main research direction.
CRediT authorship contribution statement
Xilin Luo: Software, Methodology, Visualization, Writing - original draft, Writing - review & editing, Validation, Data curation. Huiming Duan: Conceptualization, Methodology, Funding acquisition, Project administration, Resources, Supervision. Kai Xu: Investigation, Formal analysis, Validation, Data curation.
Declaration of Competing Interest
The authors declare that there is no conflict of interests regarding the publication of this paper
Acknowledgments
The authors are grateful to the editor for their valuable comments. This work is supported by the Project of Humanities and Social Sciences Planning Fund of Ministry of Education of China (18YJA630022), National Natural Science Foundation of China (71871174), Chongqing Science and Technology Foundation (cstc2020jcyj-msxmX0649).
Appendix
See Table 12, Table 13, Table 14, Table 15, Table 16.
Table 12.
Results of three grey models in Validation Case 1.
| Raw data | GM (1,1) | APE (%) | GRM (1,1) | APE (%) | GRM (1,1,) | APE (%) | Raw data | GM (1,1) | APE (%) | GRM (1,1) | APE (%) | GERM (1,1,) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 10.0000 | 0.0000 | 10.0000 | 0.0000 | 10.0000 | 0.0000 | 40 | 37.7643 | -5.5893 | 39.7978 | -0.5055 | 39.8225 | -0.4439 |
| 11 | 18.9264 | 23.2912 | 9.2933 | -3.073 | 10.2359 | -0.304 | 41 | 38.6747 | -5.6716 | 40.7426 | -0.6279 | 40.8005 | -0.4865 |
| 12 | 19.3826 | 2.0644 | 11.0746 | -2.3283 | 11.3413 | 0.0629 | 42 | 39.607 | -5.6976 | 41.6918 | -0.7339 | 41.779 | -0.5262 |
| 13 | 19.8499 | -4.9464 | 12.6151 | -2.0157 | 12.4385 | 0.1018 | 43 | 40.5618 | -5.6702 | 42.6457 | -0.8239 | 42.7581 | -0.5625 |
| 14 | 20.3284 | -5.5739 | 14.0047 | -1.7336 | 13.5264 | 0.1188 | 44 | 41.5396 | -5.5918 | 43.6048 | -0.8982 | 43.7382 | -0.5949 |
| 15 | 20.8185 | -2.2889 | 15.2903 | -1.3993 | 14.6053 | 0.125 | 45 | 42.541 | -5.4644 | 44.5694 | -0.9569 | 44.7195 | -0.6233 |
| 16 | 21.3203 | 3.9993 | 16.4999 | -0.9970 | 15.6757 | 0.1249 | 46 | 43.5665 | -5.2901 | 45.5397 | -1.0006 | 45.7023 | -0.6471 |
| 17 | 21.8343 | 12.9983 | 17.6516 | -0.5280 | 16.7382 | 0.1209 | 47 | 44.6168 | -5.0706 | 46.5162 | -1.0295 | 46.6869 | -0.6662 |
| 18 | 22.3606 | 24.7247 | 18.7582 | 0.0021 | 17.7932 | 0.1143 | 48 | 45.6924 | -4.8076 | 47.499 | -1.0438 | 47.6735 | -0.6803 |
| 19 | 22.8997 | 39.389 | 19.8288 | 0.5874 | 18.8409 | 0.1058 | 49 | 46.7939 | -4.5023 | 48.4884 | -1.044 | 48.6623 | -0.6891 |
| 20 | 23.4517 | 17.2587 | 20.8704 | 4.3521 | 19.8819 | -0.5905 | 50 | 47.9219 | -4.1561 | 49.4848 | -1.0303 | 49.6537 | -0.6925 |
| 21 | 24.0171 | 14.3671 | 21.8884 | 4.2302 | 20.9165 | -0.3977 | 51 | 49.0772 | -3.7702 | 50.4884 | -1.0031 | 50.6480 | -0.6903 |
| 22 | 24.5961 | 11.8003 | 22.8869 | 4.0312 | 21.945 | -0.2498 | 52 | 50.2603 | -3.3456 | 51.4994 | -0.9627 | 51.6452 | -0.6823 |
| 23 | 25.1890 | 9.5174 | 23.8694 | 3.7801 | 22.9679 | -0.1394 | 53 | 51.4719 | -2.8832 | 52.5181 | -0.9092 | 52.6458 | -0.6683 |
| 24 | 25.7962 | 7.4843 | 24.8389 | 3.4953 | 23.9855 | -0.0602 | 54 | 52.7127 | -2.3838 | 53.5447 | -0.8431 | 53.6499 | -0.6483 |
| 25 | 26.4181 | 5.6724 | 25.7975 | 3.1902 | 24.9982 | -0.0072 | 55 | 53.9835 | -1.8482 | 54.5795 | -0.7646 | 54.6579 | -0.6221 |
| 26 | 27.055 | 4.0575 | 26.7475 | 2.8749 | 26.0063 | 0.0242 | 56 | 55.2849 | -1.277 | 55.6226 | -0.674 | 55.6699 | -0.5895 |
| 27 | 27.7072 | 2.6191 | 27.6904 | 2.5569 | 27.0101 | 0.0375 | 57 | 56.6176 | -0.6709 | 56.6743 | -0.5714 | 56.6862 | -0.5506 |
| 28 | 28.3751 | 1.3397 | 28.6277 | 2.2418 | 28.0101 | 0.036 | 58 | 57.9825 | -0.0302 | 57.7348 | -0.4572 | 57.7070 | -0.5051 |
| 29 | 29.0591 | 0.2039 | 29.5608 | 1.9337 | 29.0064 | 0.0222 | 59 | 59.3803 | 0.6445 | 58.8044 | -0.3315 | 58.7327 | -0.4531 |
| 30 | 29.7597 | -0.8011 | 30.4907 | 1.6358 | 29.9996 | -0.0014 | 60 | 60.8117 | 1.3529 | 59.8832 | -0.1947 | 59.7633 | -0.3944 |
| 31 | 30.4771 | -1.6868 | 31.4186 | 1.3503 | 30.9898 | -0.0328 | 61 | 62.2777 | 2.0946 | 60.9714 | -0.0469 | 60.7993 | -0.3291 |
| 32 | 31.2118 | -2.4631 | 32.3452 | 1.0788 | 31.9775 | -0.0704 | 62 | 63.7791 | 2.8694 | 62.0693 | 0.1117 | 61.8407 | -0.2569 |
| 33 | 31.9642 | -3.1387 | 33.2714 | 0.8225 | 32.9628 | -0.1126 | 63 | 65.3166 | 3.6771 | 63.1770 | 0.2809 | 62.8879 | -0.1779 |
| 34 | 32.7348 | -3.7213 | 34.198 | 0.5823 | 33.9462 | -0.1581 | 64 | 66.8912 | 4.5174 | 64.2947 | 0.4605 | 63.9411 | -0.0921 |
| 35 | 33.5239 | -4.2174 | 35.1255 | 0.3586 | 34.928 | -0.2057 | 65 | 68.5037 | 5.3903 | 65.4227 | 0.6503 | 65.0005 | 0.0007 |
| 36 | 34.3321 | -4.6331 | 36.0547 | 0.1518 | 35.9084 | -0.2545 | 66 | 70.1551 | 6.2956 | 66.5611 | 0.8501 | 66.0663 | 0.1005 |
| 37 | 35.1597 | -4.9738 | 36.986 | -0.0379 | 36.8877 | -0.3034 | 67 | 71.8463 | 7.2333 | 67.7101 | 1.0599 | 67.1388 | 0.2072 |
| 38 | 36.0073 | -5.2439 | 37.9199 | -0.2107 | 37.8663 | -0.3517 | 68 | 73.5783 | 8.2034 | 68.87 | 1.2794 | 68.2183 | 0.321 |
| 39 | 36.8753 | -5.4479 | 38.8571 | -0.3665 | 38.8445 | -0.3988 | 69 | 75.3521 | 9.2059 | 70.0408 | 1.5084 | 69.3049 | 0.4418 |
| 70 | 77.1686 | 10.2408 | 71.2229 | 1.747 | 70.3988 | 0.5698 |
Table 13.
Fitting values of models in Case 1.
| Date | Raw data | GM (1,1) | APE (%) | Verhulst | APE (%) | ARGM (1,1) | APE (%) | ONGM (1,1) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1.23 | 131 | 131.0000 | 0.0000 | 131.0000 | 0.0000 | 131 | 0.0000 | 131.0000 | 0.0000 |
| 1.24 | 261 | 525.4234 | 101.3116 | 94.8453 | -63.6608 | 465.5988 | 78.3903 | 188.401 | 27.8157 |
| 1.25 | 462 | 647.4437 | 40.1393 | 161.5834 | -65.0252 | 756.06 | 63.6494 | 489.2726 | -5.9032 |
| 1.26 | 688 | 797.8011 | 15.9595 | 271.6218 | -60.5201 | 1008.2059 | 46.5416 | 775.2303 | -12.6788 |
| 1.27 | 776 | 983.0764 | 26.6851 | 446.4831 | -42.4635 | 1227.0907 | 58.1302 | 1047.0131 | -34.9244 |
| 1.28 | 1772 | 1211.3786 | -31.6378 | 707.5696 | -60.0694 | 1417.102 | -20.0281 | 1305.3239 | 26.3361 |
| 1.29 | 1462 | 1492.7000 | 2.0999 | 1058.8516 | -27.5751 | 1582.0485 | 8.2113 | 1550.8304 | -6.0759 |
| 1.30 | 1741 | 1839.3533 | 5.6492 | 1455.9708 | -16.3716 | 1725.2366 | -0.9054 | 1784.1672 | -2.4794 |
| 1.31 | 1984 | 2266.5107 | 14.2395 | 1785.2687 | -10.0167 | 1849.5365 | -6.7774 | 2005.9377 | -1.1057 |
| 2.1 | 2101 | 2792.8680 | 32.9304 | 1905.3780 | -9.3109 | 1957.4397 | -6.8330 | 2216.7151 | 5.5076 |
| 2.2 | 2590 | 3441.4625 | 32.8750 | 1754.7511 | -32.249 | 2051.1092 | -20.8066 | 2417.0444 | -6.6778 |
| 2.3 | 2827 | 4240.6817 | 50.0064 | 1409.742 | -50.1329 | 2132.4226 | -24.5694 | 2607.4435 | -7.7664 |
| 2.4 | 3233 | 5225.5054 | 61.6302 | 1013.4274 | -68.6537 | 2203.0097 | -31.8587 | 2788.4047 | -13.7518 |
| 2.5 | 3892 | 6439.0372 | 65.4429 | 671.7559 | -82.7401 | 2264.2856 | -41.8221 | 2960.3957 | -23.9364 |
| 2.6 | 3697 | 7934.3904 | 114.617 | 421.6564 | -88.5946 | 2317.4784 | -37.3146 | 3123.8612 | -15.5028 |
| Date | Raw data | ENGM (1,1) | APE (%) | ARIMA | APE (%) | GRM (1,1) | APE (%) | GERM(1,1,) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1.23 | 131 | 131.0000 | 0.0000 | 587.2736 | -348.3005 | 131.0000 | 0.0000 | 131.0000 | 0.0000 |
| 1.24 | 261 | 107.9406 | -58.6435 | 657.3624 | -151.863 | 378.7444 | 45.1128 | 260.9996 | -0.0002 |
| 1.25 | 462 | 439.9931 | -4.7634 | 662.6214 | -43.4246 | 624.1423 | 35.0957 | 469.4866 | 1.6205 |
| 1.26 | 688 | 748.3673 | 8.7743 | 775.4638 | -12.7128 | 869.6805 | 26.4071 | 688.0390 | 0.0057 |
| 1.27 | 776 | 1034.7516 | 33.3443 | 1074.8507 | -38.5117 | 1116.2213 | 43.8429 | 918.1424 | 18.3173 |
| 1.28 | 1772 | 1300.714 | -26.5963 | 1459.0940 | 17.6584 | 1364.2258 | -23.0121 | 1160.6658 | -34.4997 |
| 1.29 | 1462 | 1547.7108 | 5.8626 | 1391.2421 | 4.8398 | 1613.9876 | 10.3959 | 1416.0809 | -3.1408 |
| 1.30 | 1741 | 1777.0945 | 2.0732 | 2069.5928 | -18.8738 | 1865.7134 | 7.1633 | 1684.6594 | -3.2361 |
| 1.31 | 1984 | 1990.121 | 0.3085 | 2345.1462 | -18.2029 | 2119.5596 | 6.8326 | 1966.6041 | -0.8768 |
| 2.1 | 2101 | 2187.9568 | 4.1388 | 2220.4564 | -5.6857 | 2375.6505 | 13.0724 | 2262.1216 | 7.6688 |
| 2.2 | 2590 | 2371.6851 | -8.4291 | 2184.6567 | 15.6503 | 2634.0888 | 1.7023 | 2571.4581 | -0.7159 |
| 2.3 | 2827 | 2542.3119 | -10.0703 | 2361.6271 | 16.4617 | 2894.9622 | 2.4040 | 2894.9135 | 2.4023 |
| 2.4 | 3233 | 2700.7714 | -16.4624 | 2993.5868 | 7.4053 | 3158.3475 | -2.3091 | 3232.8462 | -0.0048 |
| 2.5 | 3892 | 2847.9312 | -26.8260 | 3475.7700 | 10.6945 | 3424.3133 | -12.0166 | 3585.6721 | -7.8707 |
| 2.6 | 3697 | 2984.5973 | -19.2698 | 3542.2804 | 4.1850 | 3692.9217 | -0.1103 | 3953.8634 | 6.9479 |
Table 14.
Fitting values of models in Case 2.
| Date | Raw data | GM (1,1) | APE (%) | Verhulst | APE (%) | ARGM (1,1) | APE (%) | ONGM (1,1) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 3.10 | 977 | 977.0000 | 0.0000 | 977.0000 | 0.0000 | 977.0000 | 0.0000 | 977.0000 | 0.0000 |
| 3.11 | 2313 | 2295.3639 | -0.7625 | 411.9399 | -82.1902 | 1868.5143 | -19.2168 | 2551.2386 | -10.3000 |
| 3.12 | 2651 | 2518.0459 | -5.0152 | 578.7551 | -78.1684 | 2575.0435 | -2.8652 | 2658.438 | -0.2806 |
| 3.13 | 2547 | 2762.3310 | 8.4543 | 806.3475 | -68.3413 | 3134.9711 | 23.0848 | 2798.3715 | -9.8693 |
| 3.14 | 3497 | 3030.3152 | -13.3453 | 1110.4411 | -68.2459 | 3578.7163 | 2.3368 | 2981.0345 | 14.7545 |
| 3.15 | 3590 | 3324.2975 | -7.4012 | 1504.9561 | -58.0792 | 3930.3864 | 9.4815 | 3219.4750 | 10.3210 |
| 3.16 | 3233 | 3646.8002 | 12.7993 | 1996.0261 | -38.2609 | 4209.0865 | 30.1914 | 3530.7248 | -9.2089 |
| 3.17 | 3526 | 4000.5901 | 13.4597 | 2572.7846 | -27.0339 | 4429.9576 | 25.6369 | 3937.0168 | -11.6567 |
| 3.18 | 4207 | 4388.7024 | 4.3191 | 3196.8035 | -24.0123 | 4604.9990 | 9.4604 | 4467.3728 | -6.1890 |
| 3.19 | 5318 | 4814.4671 | -9.4685 | 3796.0134 | -28.6195 | 4743.7201 | -10.7988 | 5159.6764 | 2.9771 |
| 3.20 | 5986 | 5281.5367 | -11.7685 | 4272.1207 | -28.6315 | 4853.6572 | -18.9165 | 6063.3796 | 1.2927 |
| 3.21 | 6557 | 5793.9186 | -11.6377 | 4527.1791 | -30.9565 | 4940.7828 | -24.6487 | 7243.0346 | 10.4626 |
| Date | Raw data | ENGM (1,1) | APE (%) | ARIMA | APE (%) | GRM (1,1) | APE (%) | GERM (1,1,) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 3.10 | 977 | 977.0000 | 0.0000 | 2103.8538 | -115.3382 | 977.0000 | 0.0000 | 977.0000 | 0.0000 |
| 3.11 | 2313 | 2778.3139 | 20.1173 | 1753.5983 | 24.1851 | 2295.1304 | -0.7726 | 2345.6369 | 1.4110 |
| 3.12 | 2651 | 2961.7041 | 11.7203 | 2612.2324 | 1.4624 | 2517.9964 | -5.0171 | 2506.3437 | -5.4567 |
| 3.13 | 2547 | 3186.8406 | 25.1213 | 2849.9128 | -11.8929 | 2762.3603 | 8.4554 | 2669.8166 | 4.8220 |
| 3.14 | 3497 | 3463.2265 | -0.9658 | 2889.2604 | 17.3789 | 3030.3800 | -13.3434 | 2859.3663 | -18.2337 |
| 3.15 | 3590 | 3802.5278 | 5.9200 | 3512.8958 | 2.1477 | 3324.3702 | -7.3992 | 3090.2263 | -13.9213 |
| 3.16 | 3233 | 4219.0666 | 30.5001 | 3633.3998 | -12.3848 | 3646.8581 | 12.8011 | 3378.2706 | 4.4934 |
| 3.17 | 3526 | 4730.4249 | 34.1584 | 3543.4230 | -0.4941 | 4000.6122 | 13.4604 | 3742.5556 | 6.1417 |
| 3.18 | 4207 | 5358.1871 | 27.3636 | 3819.6221 | 9.2079 | 4388.6674 | 4.3182 | 4207.0690 | 0.0016 |
| 3.19 | 5318 | 6128.8509 | 15.2473 | 4281.7874 | 19.485 | 4814.3521 | -9.4706 | 4802.5537 | -9.6925 |
| 3.20 | 5986 | 7074.9461 | 19.0126 | 4964.6274 | 17.0627 | 5281.3168 | -11.7722 | 5568.7153 | -6.9710 |
| 3.21 | 6557 | 8236.4073 | 18.1915 | 4490.2532 | 31.5197 | 5793.5659 | -11.6430 | 6557.0293 | 0.0004 |
Table 15.
Fitting values of models in Case 3.
| Date | Raw data | GM (1,1) | APE (%) | Verhulst | APE (%) | ARGM (1,1) | APE (%) | ONGM (1,1) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 4.11 | 4858.0000 | 0.0000 | 4858.0000 | 0.0000 | 4858 | 0.0000 | 4858.0000 | 0.0000 | |
| 4.12 | 4313 | 3969.9498 | -7.9539 | 3021.5485 | -29.9432 | 4072.0243 | -5.5872 | 4146.0369 | 3.8712 |
| 4.13 | 3579 | 3915.9128 | 9.4136 | 3897.1178 | 8.8885 | 3892.5527 | 8.7609 | 3810.8437 | -6.4779 |
| 4.14 | 3489 | 3862.6113 | 10.7083 | 4205.6223 | 20.5395 | 3851.5718 | 10.3919 | 3731.8115 | -6.9593 |
| 4.15 | 4178 | 3810.0353 | -8.8072 | 3748.9680 | -10.2688 | 3842.2141 | -8.0370 | 3713.1771 | 11.1255 |
| 4.16 | 4326 | 3758.1749 | -13.1259 | 2813.6517 | -34.9595 | 3840.0773 | -11.2326 | 3708.7835 | -14.2676 |
| 4.17 | 5065 | 3707.0205 | -26.8110 | 1850.8859 | -63.4573 | 3839.5894 | -24.1937 | 3707.7476 | -26.7967 |
| 4.18 | 5292 | 3656.5623 | -30.9040 | 1114.2737 | -78.9442 | 3839.478 | -27.4475 | 3707.5033 | -29.9414 |
| 4.19 | 4956 | 3606.7910 | -27.2237 | 635.3629 | -87.1799 | 3839.4526 | -22.5292 | 3707.4457 | -25.1928 |
| 4.20 | 4721 | 3557.6971 | -24.6410 | 351.1143 | -92.5627 | 3839.4468 | -18.6730 | 3707.4322 | -21.4693 |
| 4.21 | 3853 | 3509.2714 | -8.9211 | 190.6801 | -95.0511 | 3839.4454 | -0.3518 | 3707.4290 | -3.7781 |
| 4.22 | 4854 | 3461.5049 | -28.6876 | 102.5734 | -97.8868 | 3839.4451 | -20.9014 | 3707.4282 | -23.6212 |
| 4.23 | 4760 | 3414.3886 | -28.2691 | 54.8958 | -98.8467 | 3839.4451 | -19.3394 | 3707.4281 | -22.1129 |
| 4.24 | 5487 | 3367.9136 | -38.6201 | 29.2989 | -99.4660 | 3839.445 | -30.0265 | 3707.4282 | -32.4325 |
| 4.25 | 5158 | 3322.0712 | -35.5938 | 15.6144 | -99.6973 | 3839.445 | -25.5633 | 3707.4283 | -28.1228 |
| Date | Raw data | ENGM (1,1) | APE (%) | ARIMA | APE (%) | GRM (1,1) | APE (%) | GERM(1,1,) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 4.11 | 4858 | 4858.0000 | 0.0000 | 4965.9742 | -2.2226 | 4858.0000 | 0.0000 | 4858.0000 | 0.0000 |
| 4.12 | 4313 | 3998.2368 | -7.2980 | 4717.4428 | -9.3773 | 3969.8426 | -7.9564 | 4313.0979 | 0.0023 |
| 4.13 | 3579 | 3722.3409 | 4.0051 | 4339.0877 | -21.2374 | 3915.9602 | 9.4149 | 3692.1102 | 3.1604 |
| 4.14 | 3489 | 3552.3716 | 1.8163 | 3836.821 | -9.9691 | 3862.6518 | 10.7094 | 3807.9261 | 9.1409 |
| 4.15 | 4178 | 3447.6599 | -17.4806 | 4038.7789 | 3.3322 | 3809.991 | -8.8083 | 3940.0159 | -5.6961 |
| 4.16 | 4326 | 3383.1509 | -21.7949 | 4667.2344 | -7.8880 | 3758.0015 | -13.1299 | 4071.6967 | -5.8785 |
| 4.17 | 5065 | 3343.4092 | -33.9899 | 5150.3372 | -1.6848 | 3706.6903 | -26.8176 | 4202.7668 | -17.0234 |
| 4.18 | 5292 | 3318.9259 | -37.2841 | 5034.3642 | 4.8684 | 3656.0579 | -30.9135 | 4333.3862 | -18.1144 |
| 4.19 | 4956 | 3303.8426 | -33.3365 | 5216.7338 | -5.2610 | 3606.1007 | -27.2377 | 4463.7041 | -9.9333 |
| 4.20 | 4721 | 3294.5504 | -30.2150 | 5250.5274 | -11.2164 | 3556.8135 | -24.6597 | 4593.8537 | -2.6932 |
| 4.21 | 3853 | 3288.8257 | -14.6425 | 5358.3126 | -39.0686 | 3508.1901 | -8.9491 | 4723.9543 | 22.6046 |
| 4.22 | 4854 | 3285.2990 | -32.3177 | 5429.2496 | -11.8510 | 3460.2232 | -28.714 | 4854.1137 | 0.0023 |
| 4.23 | 4760 | 3283.1263 | -31.0268 | 5518.5372 | -15.9357 | 3412.9056 | -28.3003 | 4984.4298 | 4.7149 |
| 4.24 | 5487 | 3281.7878 | -40.1898 | 5598.6861 | -2.0355 | 3366.2295 | -38.6508 | 5114.9921 | -6.7798 |
| 4.25 | 5158 | 3280.9632 | -36.3908 | 5683.3861 | -10.1858 | 3320.1870 | -35.6303 | 5245.8824 | 1.7038 |
Table 16.
Fitting values of models in Case 4.
| Date | Raw data | GM (1,1) | APE (%) | Verhulst | APE (%) | ARGM (1,1) | APE (%) | ONGM (1,1) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 6.1 | 9035 | 9035.0000 | 0.0000 | 9035.0000 | 0.0000 | 9035.0000 | 0.0000 | 9035.0000 | 0.0000 |
| 6.2 | 8863 | 9094.4356 | 2.6113 | 654.3997 | -92.6165 | 8939.7883 | 0.8664 | 9223.4489 | -4.0669 |
| 6.3 | 8536 | 9014.1475 | 5.6015 | 700.7504 | -91.7906 | 8846.3892 | 3.6362 | 9127.1391 | -6.9252 |
| 6.4 | 8831 | 8934.5682 | 1.1728 | 750.2275 | -91.5046 | 8754.7682 | -0.8632 | 9032.3510 | -2.2800 |
| 6.5 | 8726 | 8855.6914 | 1.4863 | 803.0186 | -90.7974 | 8664.8914 | -0.7003 | 8939.0605 | -2.4417 |
| 6.6 | 8855 | 8777.511 | -0.8751 | 859.3189 | -90.2957 | 8576.7257 | -3.1426 | 8847.2440 | 0.0876 |
| 6.7 | 8984 | 8700.0208 | -3.1609 | 919.3312 | -89.7670 | 8490.2384 | -5.4960 | 8756.8781 | 2.5281 |
| 6.8 | 8985 | 8623.2147 | -4.0265 | 983.2653 | -89.0566 | 8405.3976 | -6.4508 | 8667.9399 | 3.5288 |
| 6.9 | 8595 | 8547.0867 | -0.5575 | 1051.3379 | -87.768 | 8322.1720 | -3.1743 | 8580.4070 | 0.1698 |
| 6.10 | 8404 | 8471.6307 | 0.8047 | 1123.7713 | -86.6281 | 8240.5309 | -1.9451 | 8494.2570 | -1.0740 |
| 6.11 | 8779 | 8396.8409 | -4.3531 | 1200.7932 | -86.3220 | 8160.4439 | -7.0459 | 8409.4682 | 4.2093 |
| 6.12 | 8987 | 8322.7113 | -7.3917 | 1282.6355 | -85.7279 | 8081.8817 | -10.0714 | 8326.0190 | 7.3549 |
| 6.13 | 8706 | 8249.2362 | -5.2465 | 1369.5327 | -84.2691 | 8004.8151 | -8.0540 | 8243.8882 | 5.3080 |
| 6.14 | 8835 | 8176.4098 | -7.4543 | 1461.7209 | -83.4553 | 7929.2156 | -10.2522 | 8163.0551 | 7.6055 |
| 6.15 | 8246 | 8104.2262 | -1.7193 | 1559.4357 | -81.0886 | 7855.0554 | -4.7410 | 8083.4991 | 1.9707 |
| 6.16 | 8248 | 8032.6800 | -2.6106 | 1662.9105 | -79.8386 | 7782.3070 | -5.6461 | 8005.2001 | 2.9437 |
| 6.17 | 7843 | 7961.7653 | 1.5143 | 1772.3734 | -77.4018 | 7710.9436 | -1.6837 | 7928.1381 | -1.0855 |
| 6.18 | 7790 | 7891.4767 | 1.3027 | 1888.0453 | -75.7632 | 7640.9387 | -1.9135 | 7852.2937 | -0.7997 |
| 6.19 | 7972 | 7821.8087 | -1.8840 | 2010.1361 | -74.7850 | 7572.2666 | -5.0142 | 7777.6476 | 2.4379 |
| 6.20 | 7889 | 7752.7557 | -1.7270 | 2138.8415 | -72.8883 | 7504.9019 | -4.8688 | 7704.1809 | 2.3427 |
| 6.21 | 7728 | 7684.3123 | -0.5653 | 2274.339 | -70.5701 | 7438.8196 | -3.7420 | 7631.8750 | 1.2439 |
| 6.22 | 7600 | 7616.4731 | 0.2168 | 2416.7834 | -68.2002 | 7373.9953 | -2.9737 | 7560.7114 | 0.5170 |
| 6.23 | 7425 | 7549.2329 | 1.6732 | 2566.3021 | -65.4370 | 7310.4052 | -1.5434 | 7490.6722 | -0.8845 |
| 6.24 | 7176 | 7482.5862 | 4.2724 | 2722.9899 | -62.0542 | 7248.0256 | 1.0037 | 7421.7396 | -3.4245 |
| 6.25 | 7113 | 7416.5280 | 4.2672 | 2886.9034 | -59.4137 | 7186.8336 | 1.0380 | 7353.8961 | -3.3867 |
| 6.26 | 6800 | 7351.0529 | 8.1037 | 3058.0546 | -55.0286 | 7126.8066 | 4.8060 | 7287.1245 | -7.1636 |
| 6.27 | 6852 | 7286.1558 | 6.3362 | 3236.4050 | -52.7670 | 7067.9223 | 3.1512 | 7221.4078 | -5.3912 |
| 6.28 | 6791 | 7221.8317 | 6.3442 | 3421.8589 | -49.6119 | 7010.1591 | 3.2272 | 7156.7294 | -5.3855 |
| 6.29 | 6719 | 7158.0754 | 6.5348 | 3614.2566 | -46.2084 | 6953.4955 | 3.4900 | 7093.0730 | -5.5674 |
| 6.30 | 6693 | 7094.8820 | 6.0045 | 3813.3674 | -43.0245 | 6897.9106 | 3.0616 | 7030.4223 | -5.0414 |
| 7.1 | 6556 | 7032.2465 | 7.2643 | 4018.8832 | -38.6992 | 6843.3840 | 4.3835 | 6968.7614 | -6.2959 |
| 7.2 | 6760 | 6970.1640 | 3.1089 | 4230.4116 | -37.4199 | 6789.8954 | 0.4422 | 6908.0748 | -2.1905 |
| 7.3 | 6718 | 6908.6295 | 2.8376 | 4447.4702 | -33.7977 | 6737.4251 | 0.2892 | 6848.3469 | -1.9403 |
| 7.4 | 6632 | 6847.6383 | 3.2515 | 4669.4810 | -29.5917 | 6685.9538 | 0.8135 | 6789.5628 | -2.3758 |
| 7.5 | 6736 | 6787.1855 | 0.7599 | 4895.7661 | -27.3194 | 6635.4623 | -1.4925 | 6731.7074 | 0.0637 |
| 7.6 | 6611 | 6727.2664 | 1.7587 | 5125.5437 | -22.4695 | 6585.9320 | -0.3792 | 6674.7661 | -0.9645 |
| 7.7 | 6368 | 6667.8763 | 4.7091 | 5357.9271 | -15.8617 | 6537.3447 | 2.6593 | 6618.7245 | -3.9373 |
| 7.8 | 6562 | 6609.0105 | 0.7164 | 5591.9235 | -14.7832 | 6489.6824 | -1.1021 | 6563.5682 | -0.0239 |
| 7.9 | 6509 | 6550.6644 | 0.6401 | 5826.436 | -10.4865 | 6442.9275 | -1.0151 | 6509.2835 | -0.0044 |
| 7.10 | 6635 | 6492.8334 | -2.1427 | 6060.2677 | -8.6621 | 6397.0626 | -3.5861 | 6455.8564 | 2.7000 |
| 7.11 | 6611 | 6435.5129 | -2.6545 | 6292.1274 | -4.8234 | 6352.0709 | -3.9166 | 6403.2734 | 3.1421 |
| 7.12 | 6615 | 6378.6985 | -3.5722 | 6520.6389 | -1.4265 | 6307.9358 | -4.6419 | 6351.5213 | 3.983 |
| 7.13 | 6537 | 6322.3857 | -3.2831 | 6744.3521 | 3.1720 | 6264.6409 | -4.1664 | 6300.5867 | 3.6165 |
| 7.14 | 6248 | 6266.5700 | 0.2972 | 6961.7574 | 11.4238 | 6222.1702 | -0.4134 | 6250.457 | -0.0393 |
| 7.15 | 6422 | 6211.2470 | -3.2817 | 7171.3023 | 11.6677 | 6180.508 | -3.7604 | 6201.1192 | 3.4394 |
| 7.16 | 6428 | 6156.4125 | -4.2251 | 7371.4112 | 14.6766 | 6139.6390 | -4.4860 | 6152.5610 | 4.2850 |
| 7.17 | 6406 | 6102.0620 | -4.7446 | 7560.5062 | 18.0223 | 6099.5481 | -4.7838 | 6104.7700 | 4.7023 |
| 7.18 | 6234 | 6048.1914 | -2.9806 | 7737.0309 | 24.1102 | 6060.2203 | -2.7876 | 6057.734 | 2.8275 |
| 7.19 | 6109 | 5994.7963 | -1.8694 | 7899.4753 | 29.3088 | 6021.6413 | -1.4300 | 6011.4413 | 1.5970 |
| 7.20 | 5940 | 5941.8727 | 0.0315 | 8046.4004 | 35.4613 | 5983.7968 | 0.7373 | 5965.8799 | -0.4357 |
| 7.21 | 5842 | 5889.4162 | 0.8116 | 8176.4646 | 39.9600 | 5946.6727 | 1.7917 | 5921.0383 | -1.3529 |
| 7.22 | 5862 | 5837.4229 | -0.4193 | 8288.4482 | 41.3928 | 5910.2553 | 0.8232 | 5876.9053 | -0.2543 |
| 7.23 | 5848 | 5785.8886 | -1.0621 | 8381.2769 | 43.3187 | 5874.5313 | 0.4537 | 5833.4695 | 0.2485 |
| 7.24 | 5811 | 5734.8092 | -1.3111 | 8454.0434 | 45.4835 | 5839.4874 | 0.4902 | 5790.7200 | 0.3490 |
| 7.25 | 5871 | 5684.1808 | -3.1821 | 8506.0257 | 44.8821 | 5805.1106 | -1.1223 | 5748.6459 | 2.0840 |
| 7.26 | 5765 | 5633.9993 | -2.2723 | 8536.7023 | 48.0781 | 5771.3883 | 0.1108 | 5707.2366 | 1.0020 |
| 7.27 | 5635 | 5584.2609 | -0.9004 | 8545.7630 | 51.6551 | 5738.3079 | 1.8333 | 5666.4815 | -0.5587 |
| 7.28 | 5395 | 5534.9615 | 2.5943 | 8533.1161 | 58.1671 | 5705.8574 | 5.7620 | 5626.3703 | -4.2886 |
| 7.29 | 5475 | 5486.0974 | 0.2027 | 8498.8897 | 55.2309 | 5674.0246 | 3.6352 | 5586.8929 | -2.0437 |
| 7.30 | 5509 | 5437.6647 | -1.2949 | 8443.4295 | 53.2661 | 5642.7978 | 2.4287 | 5548.0392 | -0.7086 |
| 7.31 | 5482 | 5389.6595 | -1.6844 | 8367.2909 | 52.6321 | 5612.1655 | 2.3744 | 5509.7993 | -0.5071 |
| 8.1 | 5462 | 5342.0782 | -2.1956 | 8271.2278 | 51.4322 | 5582.1164 | 2.1991 | 5472.1637 | 0.1861 |
| 8.2 | 5427 | 5294.9169 | -2.4338 | 8156.1765 | 50.2889 | 5552.6393 | 2.3151 | 5435.1226 | 0.1497 |
| 8.3 | 5394 | 5248.1719 | -2.7035 | 8023.2368 | 48.7437 | 5523.7235 | 2.4050 | 5398.6668 | 0.0865 |
| 8.4 | 5159 | 5201.8397 | 0.8304 | 7873.6503 | 52.6197 | 5495.3581 | 6.5198 | 5362.787 | 3.9501 |
| 8.5 | 5204 | 5155.9165 | -0.9240 | 7708.7764 | 48.1318 | 5467.5327 | 5.0640 | 5327.4741 | 2.3727 |
| 8.6 | 5267 | 5110.3986 | -2.9733 | 7530.0675 | 42.9669 | 5440.2370 | 3.2891 | 5292.7191 | 0.4883 |
| 8.7 | 5241 | 5065.2827 | -3.3527 | 7339.0432 | 40.0314 | 5413.4610 | 3.2906 | 5258.5132 | 0.3342 |
| 8.8 | 5212 | 5020.5650 | -3.6730 | 7137.2651 | 36.9391 | 5387.1948 | 3.3614 | 5224.8478 | 0.2465 |
| 8.9 | 5189 | 4976.2421 | -4.1002 | 6926.3122 | 33.4807 | 5361.4286 | 3.3230 | 5191.7142 | 0.0523 |
| 8.10 | 5118 | 4932.3105 | -3.6282 | 6707.7578 | 31.0621 | 5336.1529 | 4.2625 | 5159.1042 | 0.8031 |
| 8.11 | 4945 | 4888.7668 | -1.1372 | 6483.1485 | 31.1051 | 5311.3584 | 7.4087 | 5127.0094 | 3.6807 |
| 8.12 | 5102 | 4845.6075 | -5.0253 | 6253.9852 | 22.5791 | 5287.0359 | 3.6267 | 5095.4216 | -0.1289 |
| (continued on next page) | |||||||||
Table 16.
(continued)
| Date | Raw data | ENGM (1,1) | APE (%) | ARIMA | APE (%) | GRM (1,1) | APE (%) | GERM(1,1,) | APE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 6.1 | 9035 | 9035.0000 | 0.0000 | 9035.0000 | 0.0000 | 9035.0000 | 0.0000 | 9035.0000 | 0.0000 |
| 6.2 | 8863 | 9234.8451 | 4.1955 | 8638.5028 | 2.5330 | 9089.6062 | 2.5568 | 9191.9322 | 3.7113 |
| 6.3 | 8536 | 9133.6364 | 7.0014 | 8556.2779 | -0.2376 | 9011.0582 | 5.5653 | 9111.4905 | 6.7419 |
| 6.4 | 8831 | 9034.2442 | 2.3015 | 8606.9425 | 2.5372 | 8932.5335 | 1.1497 | 9024.1491 | 2.1872 |
| 6.5 | 8726 | 8936.6359 | 2.4139 | 8902.1140 | -2.0183 | 8854.3859 | 1.4713 | 8934.3398 | 2.3876 |
| 6.6 | 8855 | 8840.7795 | -0.1606 | 8995.2505 | -1.5839 | 8776.7431 | -0.8838 | 8843.8336 | -0.1261 |
| 6.7 | 8984 | 8746.6435 | -2.642 | 8955.4222 | 0.3181 | 8699.6642 | -3.1649 | 8753.5069 | -2.5656 |
| 6.8 | 8985 | 8654.1971 | -3.6817 | 8802.5761 | 2.0303 | 8623.1798 | -4.0269 | 8663.8463 | -3.5743 |
| 6.9 | 8595 | 8563.4098 | -0.3675 | 8657.6387 | -0.7288 | 8547.3069 | -0.5549 | 8575.1412 | -0.2311 |
| 6.10 | 8404 | 8474.2521 | 0.8359 | 8518.8256 | -1.3663 | 8472.0546 | 0.8098 | 8487.5701 | 0.9944 |
| 6.11 | 8779 | 8386.6945 | -4.4687 | 8514.8579 | 3.0088 | 8397.4278 | -4.3464 | 8401.2449 | -4.3029 |
| 6.12 | 8987 | 8300.7084 | -7.6365 | 8709.5847 | 3.0869 | 8323.4283 | -7.3837 | 8316.2348 | -7.4637 |
| 6.13 | 8706 | 8216.2656 | -5.6253 | 8833.3304 | -1.4626 | 8250.056 | -5.2371 | 8232.5809 | -5.4378 |
| 6.14 | 8835 | 8133.3383 | -7.9418 | 8686.9944 | 1.6752 | 8177.3098 | -7.4441 | 8150.3051 | -7.7498 |
| 6.15 | 8246 | 8051.8995 | -2.3539 | 8548.7040 | -3.6709 | 8105.1874 | -1.7076 | 8069.4156 | -2.1415 |
| 6.16 | 8248 | 7971.9223 | -3.3472 | 8195.0858 | 0.6415 | 8033.686 | -2.5984 | 7989.911 | -3.1291 |
| 6.17 | 7843 | 7893.3805 | 0.6424 | 8082.0461 | -3.0479 | 7962.8022 | 1.5275 | 7911.783 | 0.8770 |
| 6.18 | 7790 | 7816.2484 | 0.3369 | 7962.3699 | -2.2127 | 7892.5324 | 1.3162 | 7835.0179 | 0.5779 |
| 6.19 | 7972 | 7740.5006 | -2.9039 | 7958.7017 | 0.1668 | 7822.8725 | -1.8706 | 7759.5986 | -2.6643 |
| 6.20 | 7889 | 7666.1124 | -2.8253 | 7978.0067 | -1.1282 | 7753.8186 | -1.7135 | 7685.505 | -2.5795 |
| 6.21 | 7728 | 7593.0593 | -1.7461 | 7831.0397 | -1.3333 | 7685.3662 | -0.5517 | 7612.715 | -1.4918 |
| 6.22 | 7600 | 7521.3173 | -1.0353 | 7573.155 | 0.3532 | 7617.511 | 0.2304 | 7541.2051 | -0.7736 |
| 6.23 | 7425 | 7450.8630 | 0.3483 | 7327.3437 | 1.3152 | 7550.2484 | 1.6868 | 7470.9509 | 0.6189 |
| 6.24 | 7176 | 7381.6732 | 2.8661 | 7165.6632 | 0.1440 | 7483.5740 | 4.2861 | 7401.9272 | 3.1484 |
| 6.25 | 7113 | 7313.7252 | 2.8219 | 7087.4427 | 0.3593 | 7417.4830 | 4.2807 | 7334.1083 | 3.1085 |
| 6.26 | 6800 | 7246.9967 | 6.5735 | 7104.7868 | -4.4822 | 7351.9709 | 8.1172 | 7267.4685 | 6.8745 |
| 6.27 | 6852 | 7181.4659 | 4.8083 | 6995.3065 | -2.0915 | 7287.0330 | 6.3490 | 7201.982 | 5.1077 |
| 6.28 | 6791 | 7117.1112 | 4.8021 | 6893.7525 | -1.5131 | 7222.6646 | 6.3564 | 7137.6229 | 5.1042 |
| 6.29 | 6719 | 7053.9116 | 4.9845 | 6729.6816 | -0.1590 | 7158.8611 | 6.5465 | 7074.3657 | 5.2890 |
| 6.30 | 6693 | 6991.8462 | 4.4651 | 6601.8851 | 1.3613 | 7095.6178 | 6.0155 | 7012.185 | 4.7689 |
| 7.1 | 6556 | 6930.8948 | 5.7183 | 6574.7506 | -0.2860 | 7032.9300 | 7.2747 | 6951.0558 | 6.0259 |
| 7.2 | 6760 | 6871.0374 | 1.6426 | 6577.7312 | 2.6963 | 6970.7930 | 3.1182 | 6890.9534 | 1.9372 |
| 7.3 | 6718 | 6812.2543 | 1.4030 | 6690.4528 | 0.4101 | 6909.2024 | 2.8461 | 6831.8536 | 1.6948 |
| 7.4 | 6632 | 6754.5262 | 1.8475 | 6703.4529 | -1.0774 | 6848.1534 | 3.2592 | 6773.7325 | 2.1371 |
| 7.5 | 6736 | 6697.8342 | -0.5666 | 6635.8891 | 1.4862 | 6787.6414 | 0.7666 | 6716.5668 | -0.2885 |
| 7.6 | 6611 | 6642.1597 | 0.4713 | 6604.6195 | 0.0965 | 6727.6619 | 1.7647 | 6660.3336 | 0.7462 |
| 7.7 | 6368 | 6587.4845 | 3.4467 | 6552.5218 | -2.8976 | 6668.2104 | 4.7144 | 6605.0105 | 3.7219 |
| 7.8 | 6562 | 6533.7905 | -0.4299 | 6501.1837 | 0.9268 | 6609.2823 | 0.7205 | 6550.5755 | -0.1741 |
| 7.9 | 6509 | 6481.0603 | -0.4292 | 6601.0316 | -1.4139 | 6550.8731 | 0.6433 | 6497.0073 | -0.1842 |
| 7.10 | 6635 | 6429.2765 | -3.1006 | 6630.2764 | 0.0712 | 6492.9784 | -2.1405 | 6444.2848 | -2.8744 |
| 7.11 | 6611 | 6378.4221 | -3.5180 | 6659.8756 | -0.7393 | 6435.5937 | -2.6532 | 6392.3876 | -3.3068 |
| 7.12 | 6615 | 6328.4804 | -4.3314 | 6594.5593 | 0.3090 | 6378.7147 | -3.5720 | 6341.2959 | -4.1376 |
| 7.13 | 6537 | 6279.4351 | -3.9401 | 6510.4546 | 0.4061 | 6322.3369 | -3.2838 | 6290.9901 | -3.7633 |
| 7.14 | 6248 | 6231.2700 | -0.2678 | 6422.2844 | -2.7894 | 6266.4561 | 0.2954 | 6241.4512 | -0.1048 |
| 7.15 | 6422 | 6183.9694 | -3.7065 | 6293.6007 | 1.9994 | 6211.0679 | -3.2845 | 6192.6608 | -3.5711 |
| 7.16 | 6428 | 6137.5178 | -4.519 | 6322.0877 | 1.6477 | 6156.1681 | -4.2289 | 6144.6008 | -4.4088 |
| 7.17 | 6406 | 6091.8998 | -4.9032 | 6331.6564 | 1.1605 | 6101.7524 | -4.7494 | 6097.2538 | -4.8196 |
| 7.18 | 6234 | 6047.1007 | -2.9981 | 6311.2151 | -1.2386 | 6047.8166 | -2.9866 | 6050.6025 | -2.9419 |
| 7.19 | 6109 | 6003.1055 | -1.7334 | 6190.6704 | -1.3369 | 5994.3565 | -1.8766 | 6004.6303 | -1.7085 |
| 7.20 | 5940 | 5959.9000 | 0.335 | 6035.7108 | -1.6113 | 5941.3680 | 0.0230 | 5959.3210 | 0.3253 |
| 7.21 | 5842 | 5917.4700 | 1.2919 | 5871.3449 | -0.5023 | 5888.8469 | 0.8019 | 5914.6589 | 1.2437 |
| 7.22 | 5862 | 5875.8014 | 0.2354 | 5772.164 | 1.5325 | 5836.7893 | -0.4301 | 5870.6284 | 0.1472 |
| 7.23 | 5848 | 5834.8808 | -0.2243 | 5765.2549 | 1.4149 | 5785.191 | -1.0740 | 5827.2148 | -0.3554 |
| 7.24 | 5811 | 5794.6946 | -0.2806 | 5772.1398 | 0.6687 | 5734.048 | -1.3242 | 5784.4034 | -0.4577 |
| 7.25 | 5871 | 5755.2296 | -1.9719 | 5736.9861 | 2.2826 | 5683.3564 | -3.1961 | 5742.1800 | -2.1942 |
| 7.26 | 5765 | 5716.473 | -0.8418 | 5681.5056 | 1.4483 | 5633.1122 | -2.2877 | 5700.5309 | -1.1183 |
| 7.27 | 5635 | 5678.4119 | 0.7704 | 5561.0200 | 1.3129 | 5583.3115 | -0.9173 | 5659.4427 | 0.4338 |
| 7.28 | 5395 | 5641.034 | 4.5604 | 5452.0088 | -1.0567 | 5533.9504 | 2.5755 | 5618.9023 | 4.1502 |
| 7.29 | 5475 | 5604.327 | 2.3621 | 5349.1247 | 2.2991 | 5485.025 | 0.1831 | 5578.8970 | 1.8977 |
| 7.30 | 5509 | 5568.2787 | 1.0760 | 5381.4724 | 2.3149 | 5436.5316 | -1.3155 | 5539.4146 | 0.5521 |
| 7.31 | 5482 | 5532.8775 | 0.9281 | 5433.6295 | 0.8824 | 5388.4663 | -1.7062 | 5500.4429 | 0.3364 |
| 8.1 | 5462 | 5498.1116 | 0.6611 | 5443.4267 | 0.3400 | 5340.8254 | -2.2185 | 5461.9703 | -0.0005 |
| 8.2 | 5427 | 5463.9697 | 0.6812 | 5294.0196 | 2.4503 | 5293.6051 | -2.4580 | 5423.9855 | -0.0555 |
| 8.3 | 5394 | 5430.4406 | 0.6756 | 5137.6747 | 4.7520 | 5246.8019 | -2.7289 | 5386.4774 | -0.1395 |
| 8.4 | 5159 | 5397.5133 | 4.6232 | 4995.8605 | 3.1622 | 5200.4119 | 0.8027 | 5349.4352 | 3.6913 |
| 8.5 | 5204 | 5365.1769 | 3.0972 | 4921.6525 | 5.4256 | 5154.4317 | -0.9525 | 5312.8486 | 2.0916 |
| 8.6 | 5267 | 5333.4209 | 1.2611 | 4903.7844 | 6.8961 | 5108.8575 | -3.0025 | 5276.7073 | 0.1843 |
| 8.7 | 5241 | 5302.2349 | 1.1684 | 4881.3277 | 6.8627 | 5063.6858 | -3.3832 | 5241.0015 | 0.0000 |
| 8.8 | 5212 | 5271.6086 | 1.1437 | 4798.975 | 7.9245 | 5018.9132 | -3.7047 | 5205.7216 | -0.1205 |
| 8.9 | 5189 | 5241.5320 | 1.0124 | 4655.2219 | 10.2867 | 4974.536 | -4.1331 | 5170.8582 | -0.3496 |
| 8.10 | 5118 | 5211.9952 | 1.8366 | 4503.7050 | 12.0026 | 4930.5507 | -3.6625 | 5136.4022 | 0.3596 |
| 8.11 | 4945 | 5182.9885 | 4.8127 | 4408.3534 | 10.8523 | 4886.9541 | -1.1738 | 5102.3448 | 3.1819 |
| 8.12 | 5102 | 5154.5024 | 1.0291 | 4391.1505 | 13.9328 | 4843.7425 | -5.0619 | 5068.6775 | -0.6531 |
References
- 1.WHO, 2020. World Health Organization. ( https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen ).
- 2.Jia W.P., Han K., Song Y. Extended SIR prediction of the epidemics trend of COVID-19 in Italy and compared with Hunan, China. Front Med. 2020;7:169. doi: 10.3389/fmed.2020.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modi S., Bhattacharya J., Basak P. Modeling and forecasting the early evolution of the Covid-19 pandemic in Brazil. ISA Trans. 2019 doi: 10.1016/j.isatra.2019.08.055. [DOI] [Google Scholar]
- 4.Yang Z., Zeng Z., Wang K. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J Thorac Dis. 2020;12:165–174. doi: 10.21037/jtd.2020.02.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomar A., Gupta N. Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Sci Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Z.X., Ge Q.Y., Li S.D. et al. Artificial intelligence forecasting of Covid-19 in China, arXiv, (2020). doi: arXiv:2002.07112.
- 7.Sina F., Amir M., Pedram G. et al. COVID-19 outbreak prediction with machine learning SSRN, 3 (2020). doi: 10.2139/ssrn.3580188.
- 8.Petropoulos F., Makridakis S. Forecasting the novel coronavirus COVID-19. PLoS One. 2020;15 doi: 10.1371/journal.pone.0231236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benvenuto D., Giovanetti, Lazzaro V.M. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data Brief. 2020;29 doi: 10.1016/j.dib.2020.105340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maleki M., Mahmoudi M.R., Wraith D. Time series modelling to forecast the confirmed and recovered cases of COVID-19. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101742. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y.F., Shou M.H., Wang Z.X. Prediction of the number of patients infected with COVID-19 based on rolling grey verhulst models. Int J Environ Res Public Health. 2020;17:4582. doi: 10.3390/ijerph17124582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo X.L., Duan H.M., He L.Y.H. A novel riccati equation grey model and its application in forecasting clean energy. Energy. 2020;205 doi: 10.1016/j.energy.2020.118085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan H.M., Lei G.Y., Shao K.L. Forecasting crude oil consumption in china using a grey prediction model with an optimal fractional-order accumulating operator. Complexity. 2018:1–12. doi: 10.1155/2018/3869619. [DOI] [Google Scholar]
- 14.Wang Z.X., Li D.D., Zheng H.H. Model comparison of GM(1,1) and DGM(1,1) based on Monte-Carlo simulation. Physica A. 2019;542 doi: 10.1016/j.physa.2019.123341. [DOI] [Google Scholar]
- 15.Richards F.J. A flexible growth function for empirical use. J Exp Bot. 1959;10:290–300. [Google Scholar]
- 16.Hsieh Y.H. Richards model: a simple procedure for real-time prediction of outbreak severity. Model Dyn Infect Dis. 2009:216–236. doi: 10.1142/9789814261265_0009. [DOI] [Google Scholar]
- 17.Hsieh Y.H., Ma S. Intervention measures, turning point, and reproduction number for Dengue, Singapore, 2005. Am J Trop Med Hyg. 2009;80:66–71. doi: 10.4269/ajtmh.2009.80.66. [DOI] [PubMed] [Google Scholar]
- 18.Chan C.H., Tuite A.R., Fisman D.N. Historical epidemiology of the second cholera pandemic: relevance to present day disease dynamics. PLoS One. 2013;8:e72498. doi: 10.1371/journal.pone.0072498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X.S., Mu J.H., Yang Y. Richards model revisited: Validation by and application to infection dynamics. J Theor Biol. 2012;313:12–19. doi: 10.1016/j.jtbi.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Yan C., Wu L.F., Liu L.Y., Kai Z. Fractional Hausdorff grey model and its properties, Chaos. Solit Fract. 2020;138 doi: 10.1016/j.chaos.2020.109915. [DOI] [Google Scholar]
- 21.Xiao X.P., Duan H.M. A new grey model for traffic flow mechanics. Eng Appl Artif Intell. 2020 doi: 10.1016/j.engappai.2019.103350. [DOI] [Google Scholar]
- 22.Duan H.M., Xiao X.P., Xiao Q.Z. An inertia grey discrete model and its application in short-term traffic flow prediction and state determination. Neural Comput Appl. 2019:1–17. doi: 10.1007/s00521-019-04364-w. [DOI] [Google Scholar]
- 23.Zeng B., Tong M.Y. and Ma X., A new structure grey Verhulst model: development and performance comparison, Appl Math Modell. 2020, (81): 522-537. doi: 10.1016/j.apm.2020.01.014.
- 24.Duan H.M., Xiao X.P. A multimode dynamic short-term traffic flow grey prediction model of high-dimension tensors. Complexity. 2019 doi: 10.1155/2019/9162163. [DOI] [Google Scholar]
- 25.Wei B., Xie N.M. On unified framework for discrete-time grey models: Extensions and applications. ISA Trans. 2020 doi: 10.1016/j.isatra.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 26.Guo X., Liu S., Wu L. Application of a novel grey self-memory coupling model to forecast the incidence rates of two notifiable diseases in China: Dysentery and Gonorrhea. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y.W., Shen Z.Z., Jiang Y. Comparison of ARIMA and GM(1,1) models for prediction of hepatitis B in China. PLoS One. 2018;13 doi: 10.1371/journal.pone.0201987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Wang L., Zheng Y. Time prediction models for echinococcosis based on gray system theory and epidemic dynamics. Int J Environ Res Public Health. 2017;14:262. doi: 10.3390/ijerph14030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S.F., Lin Y., Forrest J.Y.L. Springer; 2010. Grey systems: theory and applications. [Google Scholar]
- 30.Şahin U., Şahin T. Forecasting the cumulative number of confirmed cases of COVID-19 in Italy, UK and USA using fractional nonlinear grey Bernoulli model, Chaos. Solitons Fract. 2020;138 doi: 10.1016/j.chaos.2020.109948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng J.L. Huazhong University of Science and Technology Press; 2002. Foundations of grey theory. in Chinese. [Google Scholar]
- 32.Erdal K., Baris U., Okyay K. Grey system theory-based models in time series prediction. Expert Syst Appl. 2010;37(2):1784–1789. doi: 10.1016/j.eswa.2009.07.064. [DOI] [Google Scholar]
- 33.Wu L., Liu S., Chen H. Using a novel grey system model to forecast natural gas consumption in China. Math Probl Eng. 2015 doi: 10.1155/2015/686501. [DOI] [Google Scholar]
- 34.Chen P.Y., Yu H.M. Foundation settlement prediction based on a novel NGM model. Math Probl Eng. 2014:1–8. doi: 10.1155/2014/242809. [DOI] [Google Scholar]
- 35.Ma X., Liu Z.B. Predicting the cumulative oil field production using the novel grey ENGM model. J Comput Theor Nanosci. 2013;13(1):89–95. doi: 10.1166/jctn.2016.4773. [DOI] [Google Scholar]
- 36.Wang Q., Li S.Y., Li R.R. Will Trump's coal revival plan work? - Comparison of results based on the optimal combined forecasting technique and an extended IPAT forecasting technique. Energy. 2019;169:762–775. doi: 10.1016/j.energy.2018.12.045. [DOI] [Google Scholar]
- 37.Chen C.I., Chen H.L., Chen S.P. Forecasting of foreign exchange rates of Taiwan's major trading partners by novel nonlinear Grey Bernoulli model NGBM (1,1) Commun Nonlinear Sci Numer Simul. 2008;13:1194–1204. doi: 10.1016/j.cnsns.2006.08.008. [DOI] [Google Scholar]
- 38.Verhulst P.F. Notice sur la loi que la population poursuit dans son accroissement. Corresp Math Phys. 1838;10:113–121. [Google Scholar]
- 39.Malthus T. An essay on the principle of population, with a summary view, and introduction by professor Anthony Flew. Penguin Classics. ISBN:0-14-043206
- 40.Xiao X.P., Duan H.M., Wen J.H. A novel car-following inertia grey model and its application in forecasting short-term traffic flow. Appl Math Modell. 2020;87:546–570. doi: 10.1016/j.apm.2020.06.020. [DOI] [Google Scholar]
- 41.Zhang Q. Petroleum Industry Press; Beijing: 2002. The difference information principle of grey hazy set. [Google Scholar]
- 42.Ma X. Southwest Petroleum University; 2016. Study On Dynamical Prediction Methods Based On Grey System And Kernal Method. [Google Scholar]
- 43.Guo J.H., Li J.L., Yang X. Grey modeling for productivity prediction of low permeability oil wells. Control Dec. 2018;34:2498–2504. [Google Scholar]
- 44.Qiao Q.Z., Gao M.Y., Xiao X.P. A novel grey Riccati–Bernoulli model and its application for the clean energy consumption prediction. Eng Appl Artif Intell. 2020 doi: 10.1016/j.engappai.2020.103863. [DOI] [Google Scholar]
- 45.Zhang P., Ma X., She K. A novel power-driven grey model with whale optimization algorithm and its application in forecasting the residential energy consumption in China. Complexity. 2019:1–22. doi: 10.1155/2019/1510257. [DOI] [Google Scholar]
- 46.Hu Y.C. Electricity consumption prediction using a neural network-based grey forecasting approach. J Oper Res Soc. 2017;68:1259–1264. doi: 10.1057/s41274-016-0150-y. [DOI] [Google Scholar]
- 47.Holland J.H. Genetic algorithms. Sci Am. 1992;267(1):66–72. doi: 10.1038/scientificamerican0792-66. [DOI] [Google Scholar]