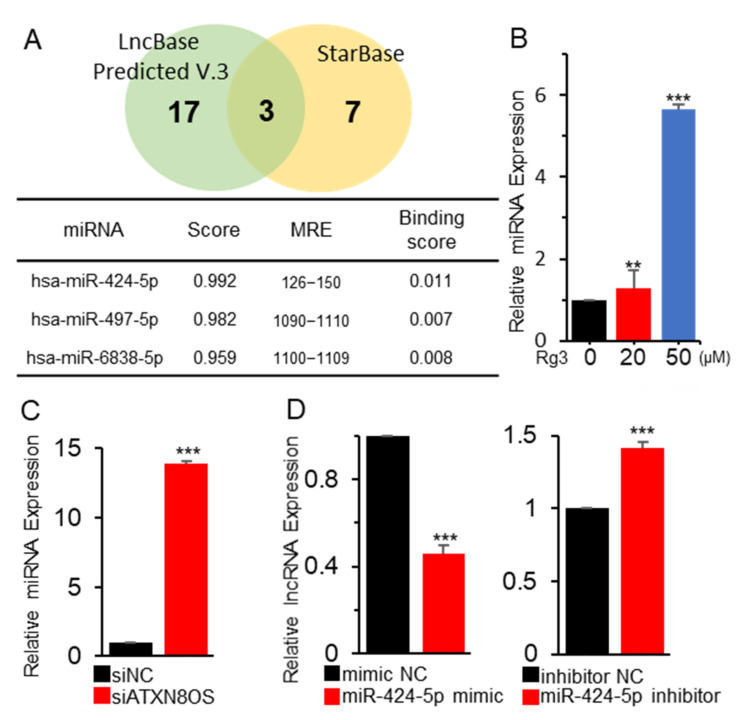
Figure 2.
ATXN8OS and miR-424-5p sponge each other. (A) Three miRs that could potentially bind ATXN8OS were screened in silico using two miR-prediction databases (LncBase Predicted v.3 and StarBase). (B) miR-424-5p exhibited the highest binding score and was therefore examined to characterize its regulation by Rg3. MCF-7 cells were treated with Rg3, and the RNA expression was quantified by qRT-PCR. (C,D) The association between the ATXN8OS and miR-424-5p expression was monitored by examining the expression of each RNA after inhibiting ATXN8OS using siRNA (C) and overexpressing (40 μM) or inhibiting miR-424-5p (20 μM) (D). All experiments were performed in triplicate, and the values are presented as the mean ± SE. Testing was done using siNC, negative control siRNA (40 μM); siATXN8OS, ATXN8OS-specific siRNA (40 μM); mimic NC, negative control mimic for miR-424-5p (40 μM); and inhibitor NC, negative control inhibitor for miR-424-5p (20 μM). ** p < 0.01, *** p < 0.001.