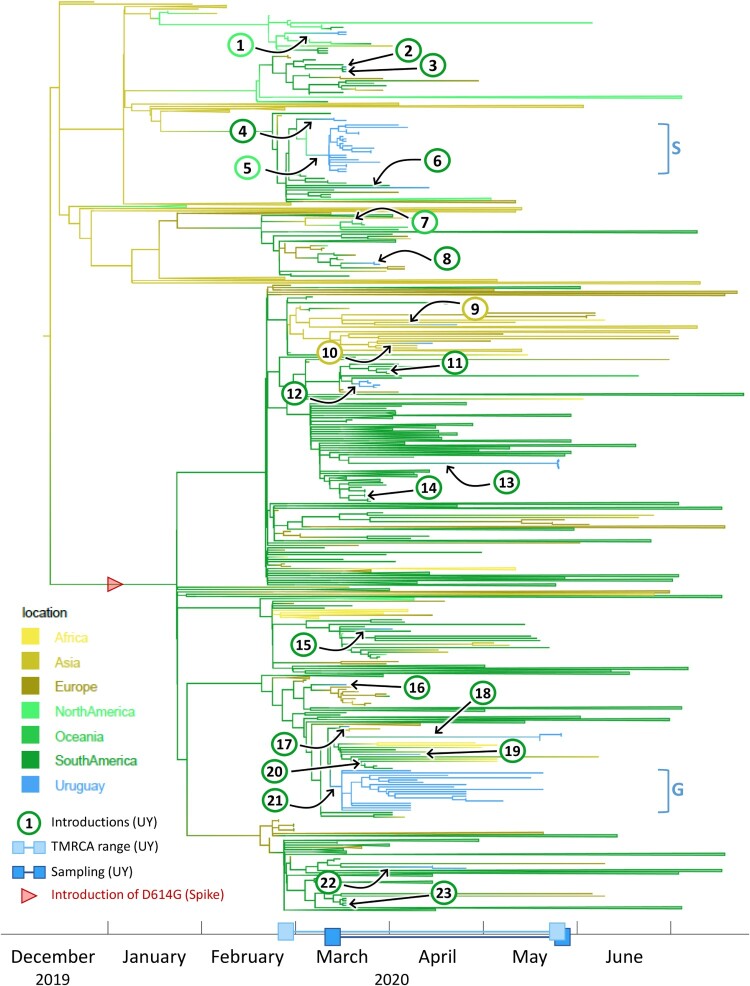
Figure 3.
Time-scaled phylogenetic tree to identify the source regions of the sequences in the imported Uruguayan clusters. We employed a discrete state phylogeography diffusion model in BEAST with seven ancestral location states (Africa, Asia, Europe, North America, Oceania, South America, and Uruguay) to identify the most probable source locations for the sequences in the 23 previously identified international introductions into Uruguay. The branches of the trees are colour-coded according to each geographic region. A colour gradient along the branches indicates historic introduction events between locations. The introductions into Uruguay are highlighted by black arrows and circles with consecutive numbering according to the introduction event (colour-code of circle outline: probable source continent). The estimated time to the most recent common ancestors (TMRCAs) of Uruguayan (UY) sequences and their sampling period are indicated as ranges along the x-axis timeline. Branches that are not involved in introduction events are collapsed to facilitate visualization. The introduction of the spike D614G mutation is indicated by a red arrowhead. The two major Uruguayan clades are highlighted by brackets, and GISAID clades are indicated.