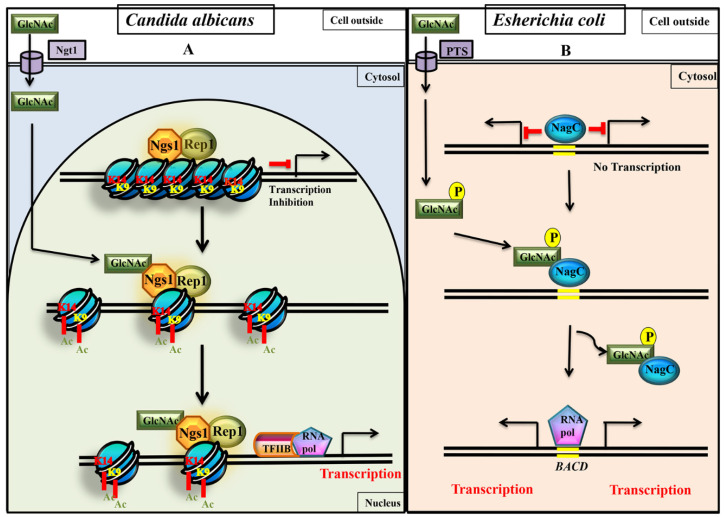
Figure 5.
Comparison of GlcNAc signaling between eukaryotes (Candida albicans) and prokaryotes (Escherichia coli). During signaling process, in C. albicans the non-phosphorylated free GlcNAc directly binds to GlcNAc sensor (Ngs1) to promote histone H3 acetylation (H3K9 Ac and H3K14 Ac) of the nucleosomes at the promoters of GlcNAc catabolic genes [34], whereas in E. coli, GlcNAc-6-phospahte formed from Phosphotransferase System (PTS) binds to NagC repressor to release it from operator region of GlcNAc catabolic genes [63,65]. (A) Working model depicting induction of GlcNAc catabolic genes through Ngs1-Rep1 mediated acetylation. More work needs to be carried out to understand the role of other proteins involved in the determination of Ngs1 specificity for GlcNAc over the other carbon sources like maltose (53). The chromatin modifying activities of activators (Ngs1-Rep1 complex) lead to altered chromatin structure (loosening of nucleosomes) thereby transcription of cognate genes is promoted. (B) In prokaryotes, as the chromatin architecture is simple, just the removal of repressor NagC is sufficient to initiate transcription.